This gallery was developed from the best images acquired from our TLL Bioimaging and Bicomputing Facility.
If you have an image that you would like to feature, please submit it to Bioimaging Group ![]() .
.
You could stand a chance to win the Image of the Month competition.
You are welcome to use these images for non-commercial purposes as long as credit is given to the person(s) who acquired the image.
- 2024
- 2021
- 2016-2020
- 2011-2015
- 2008-2010
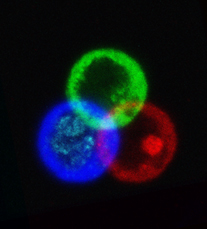
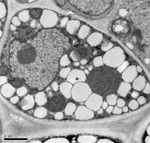
November 2024
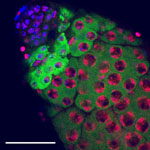
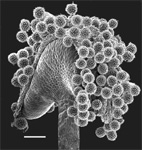
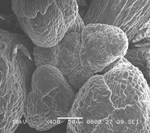
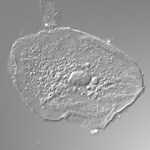
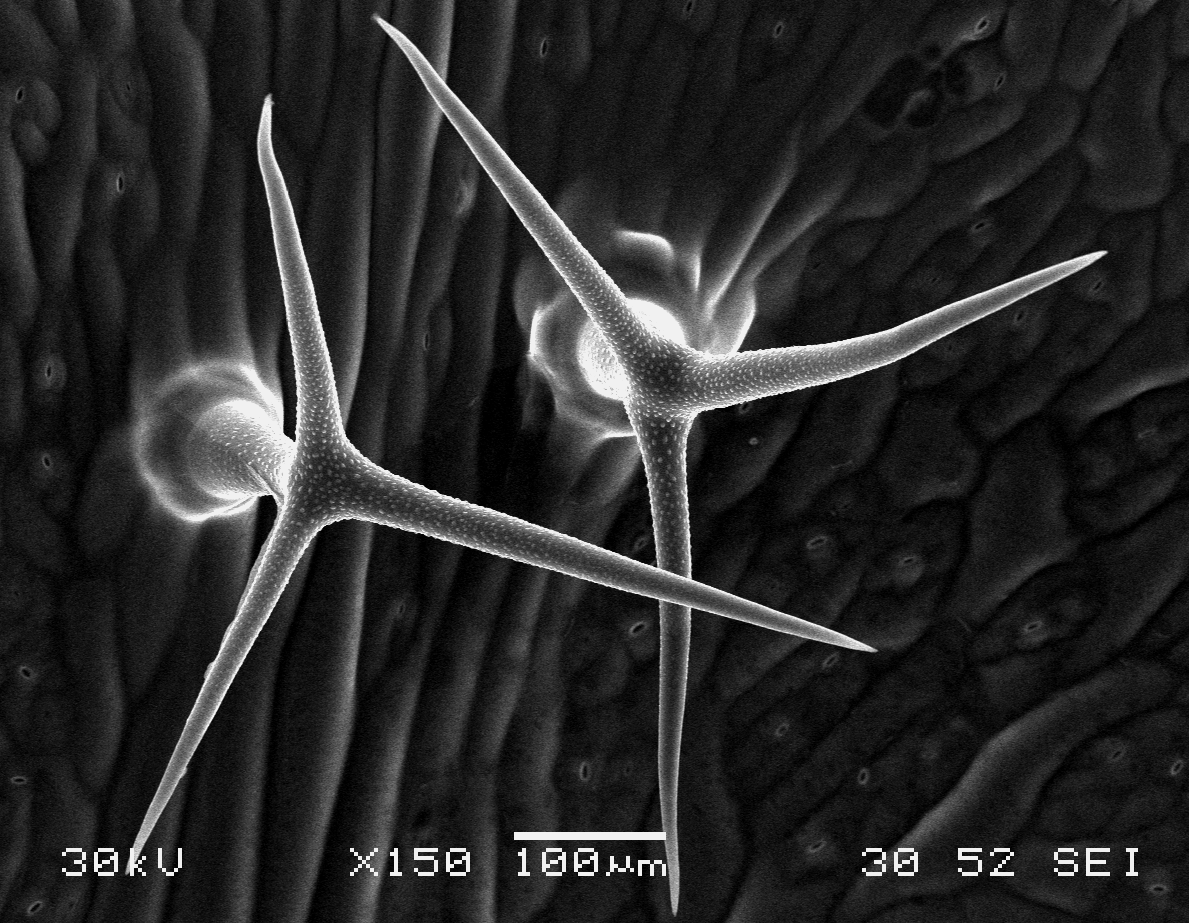
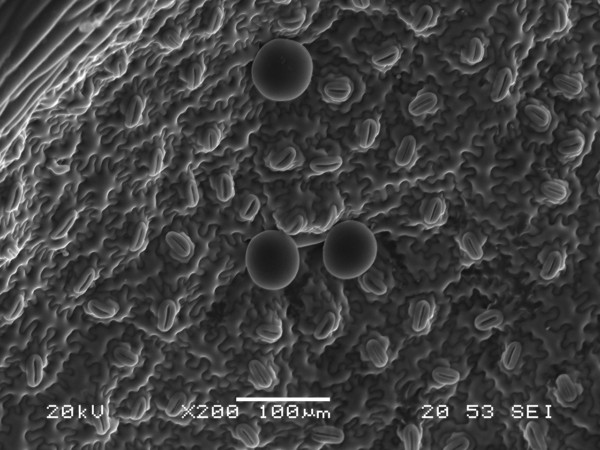
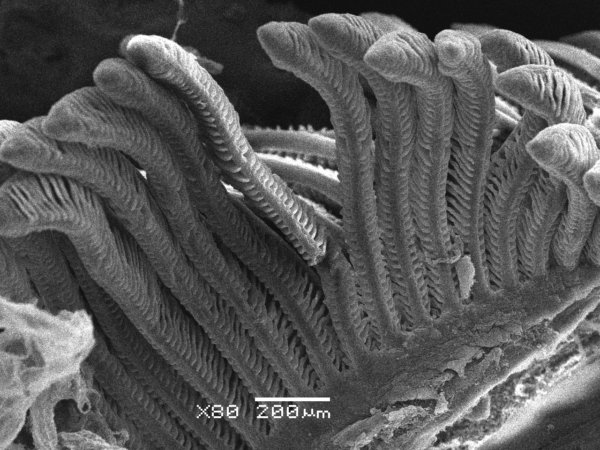
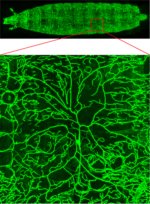
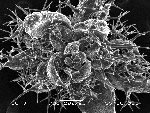
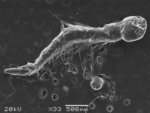
Arabidopsis trichomes (unicellular epidermis hairs) showing branched morphology
Image acquired on the JEOL JSM-6360LV Scanning Electron Microscope at 150× magnification
Taken by Jolly Madathiparambil Saju (Rajani's lab)
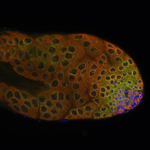
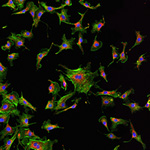
January 2024
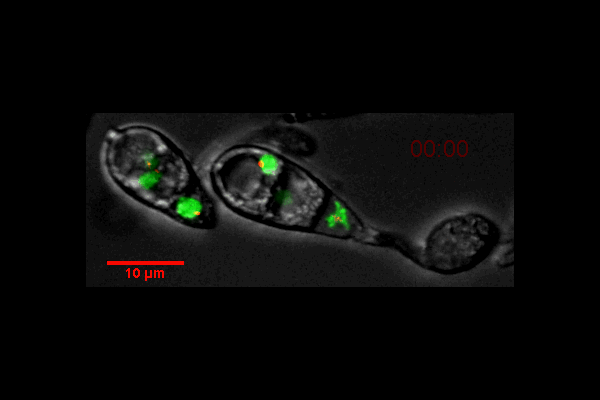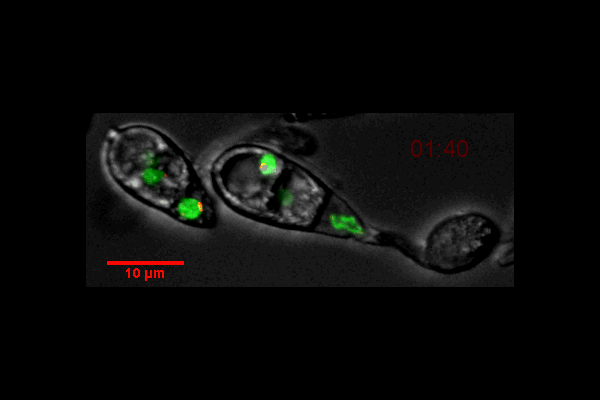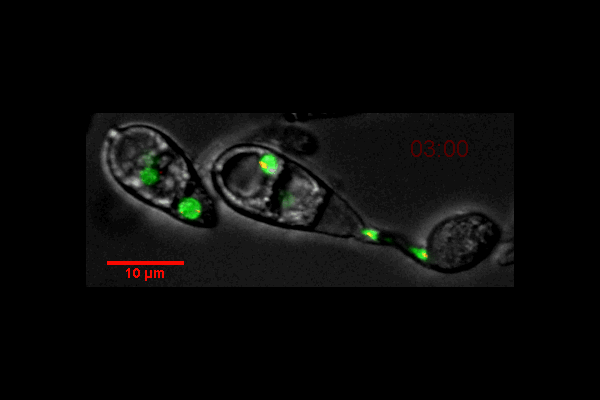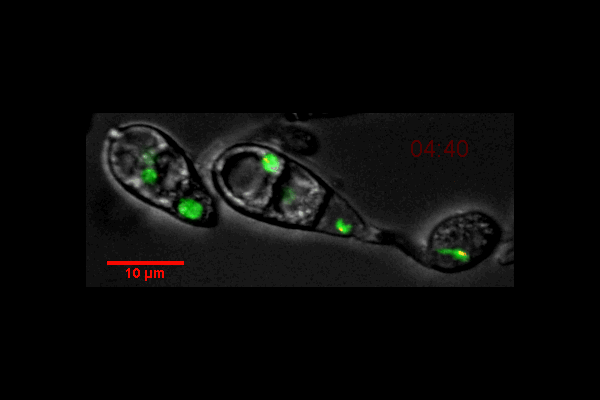
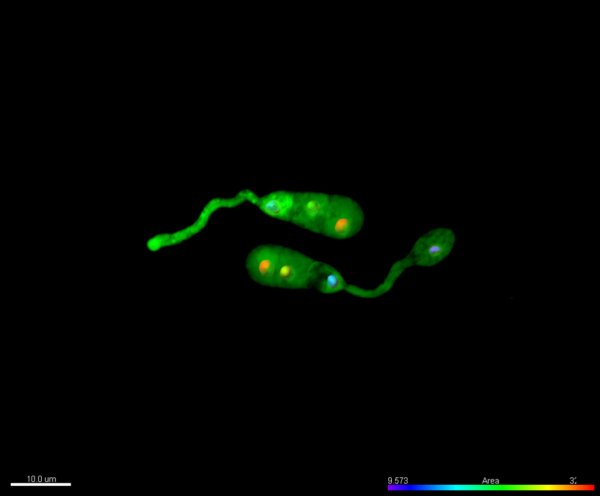
Video of Vorticella feeding
A short video clip of a Vorticella in a Cyanobacteria colony was taken at 1000x magnification showing the Vorticella feeding and snapping back.
Video was acquired using the Ocular program and then compressed using Handbrake.
Acquired using 3F Zeiss Colour Widefield System
Taken by Melvin Wong (Bioimaging Facility)
July 2021
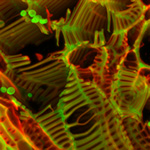
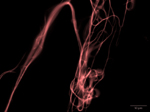
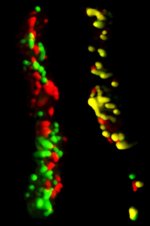
“Claw” and “Tree” - Upright stromata of Xylaria
The fungus Xylaria was cultured for over 45 days for the stromata formation. The vertical growth starts as little bumps on the culture surface and gradually form these soft bristly fibers upwards.
Acquired using Leica Fluorescence Stereomicroscope
Taken by Tham Hong Fai (Dr. Naweed Naqvi's Lab)
February 2021
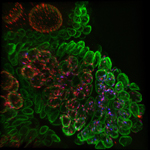
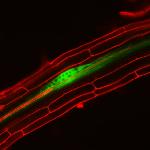
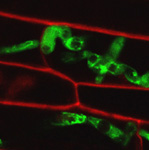
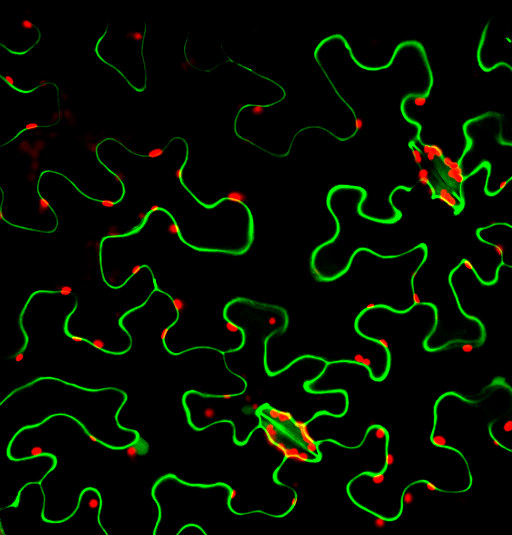
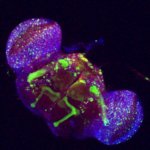
Fluorescing nanocarriers inside tobacco leaf cells
Alexa488 tagged SWNT nanocarriers showing fluorescence inside tobacco leaf cells. Red autofluorescence is seen from the chloroplasts.
Acquired using LSM5
Taken by Vaishnavi Reddy Amarr (Dr. Sarojam Rajani's Lab)
2020
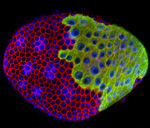
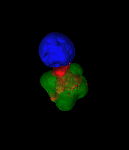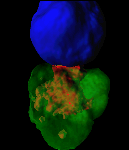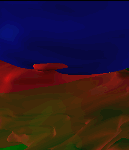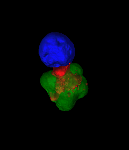
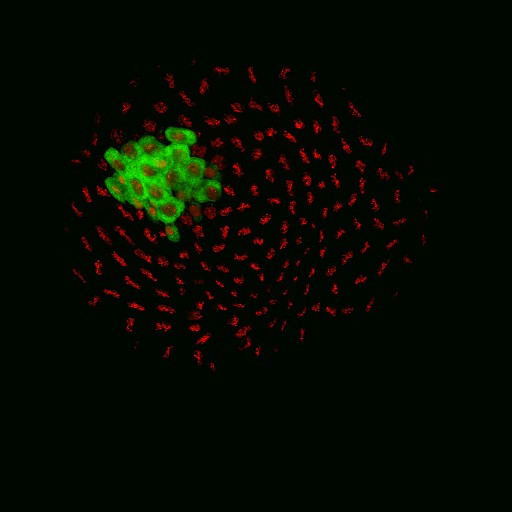
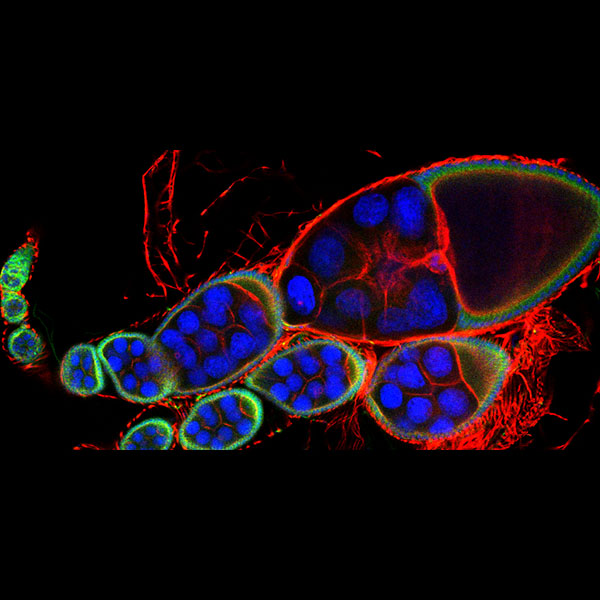
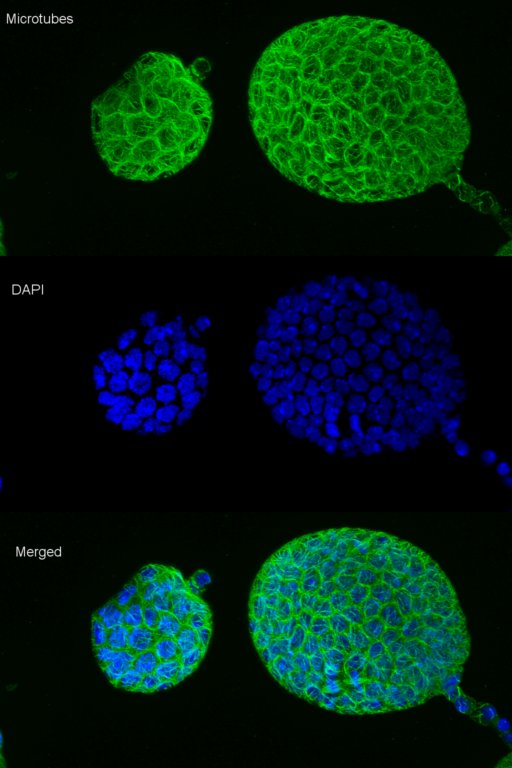
Exploring vesicle trafficking across fungus-plant interface
The surface rendered images/compartments show the vesicular sorting machinery (green, Vps35-GFP) and the biotrophy interfacial complex (red, Pwl2-mCherry.)
The image was acquired using the 100x/1.4 objective on MicroLambda Spinning Disk.
Image processing was performed using Imaris 9.5 with the new surface rendering tool.
Taken by: Zheng Wenhui (Naweed NAQVI's lab)
November 2019
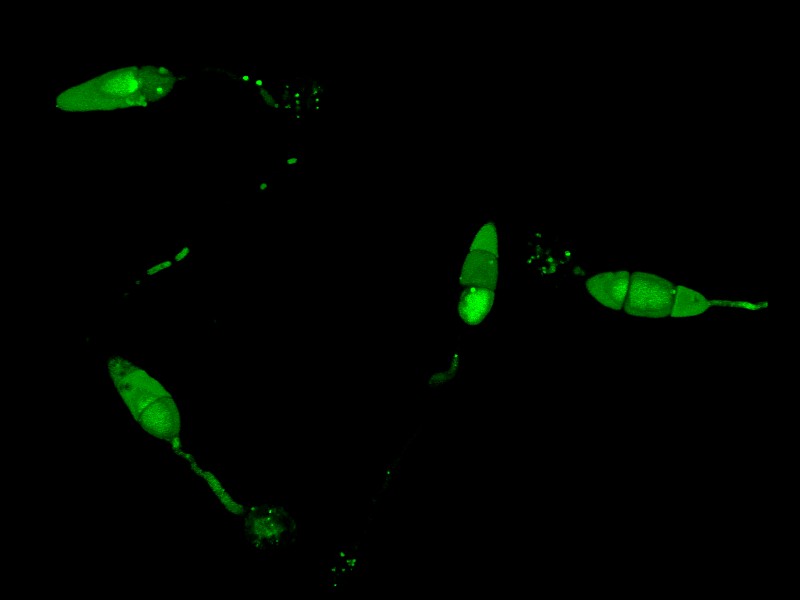
ATG8
The image shows the subcellular localization of ATG8, a key player of autophagy in the rice blast fungus Magnaporthe oryzae.
Acquired using Leica SP8, using 63x objective lens.
Taken by: Shen Qing (Naweed NAQVI's lab)
October 2019
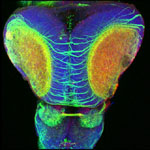
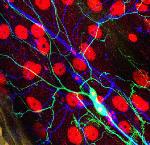
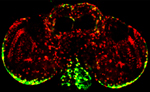
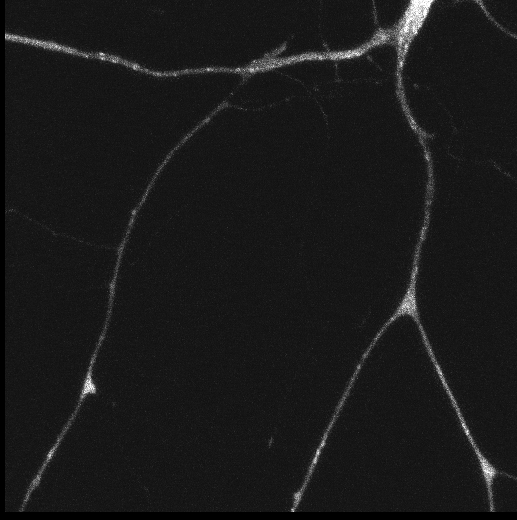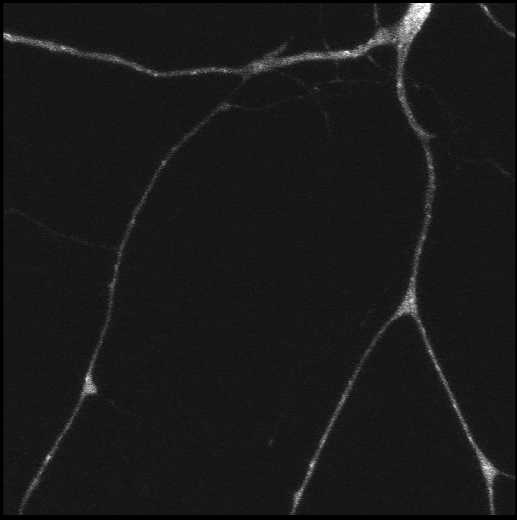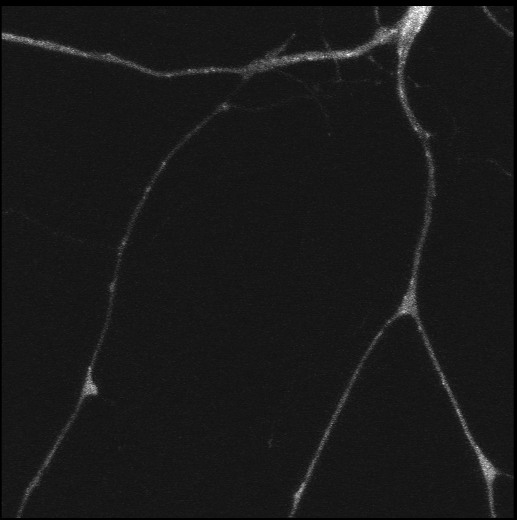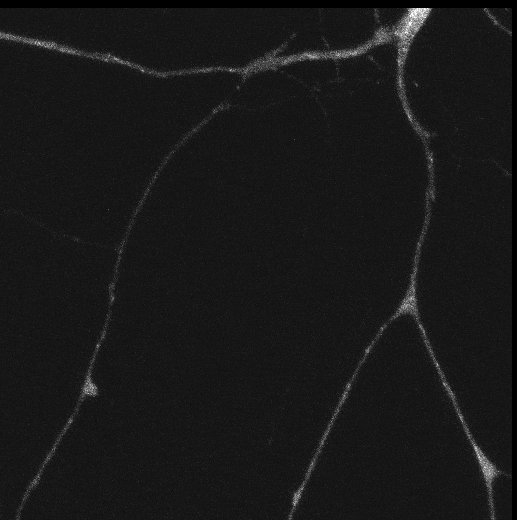
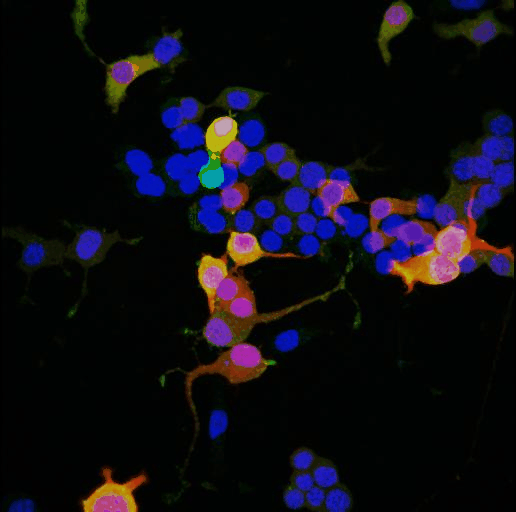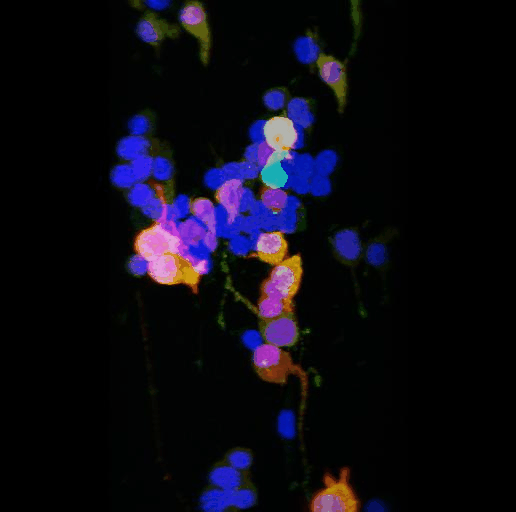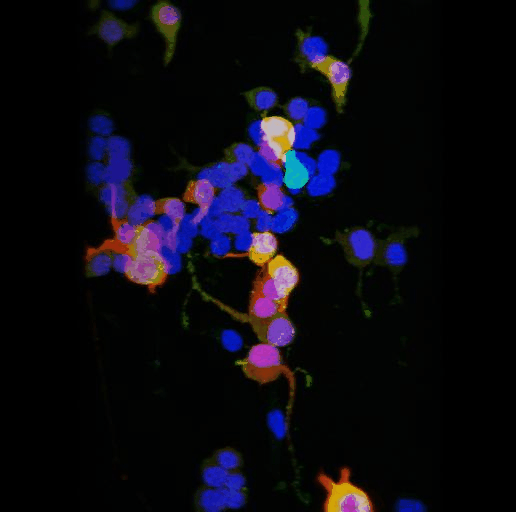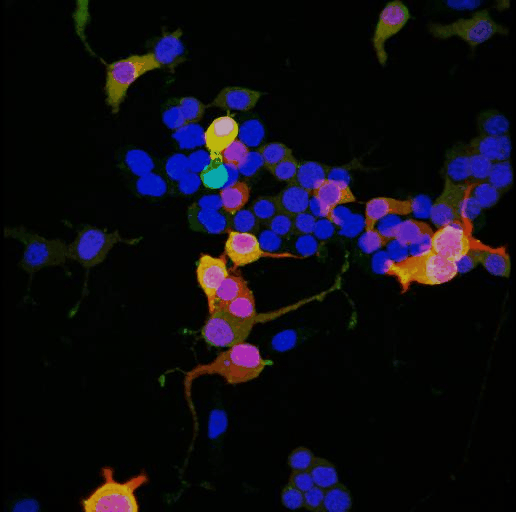
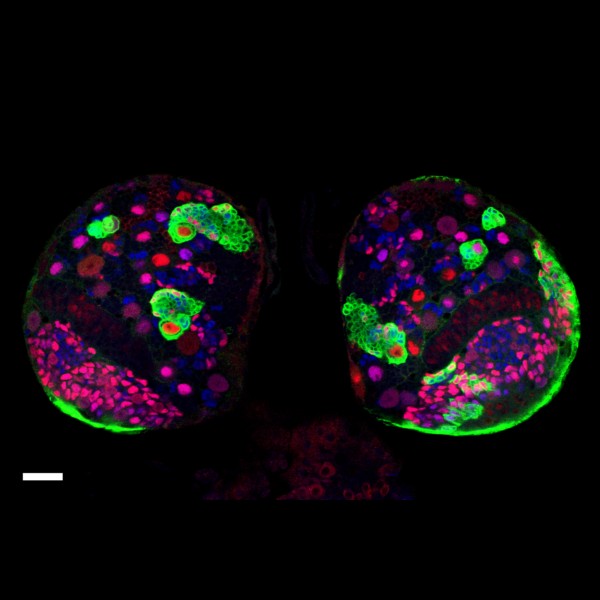
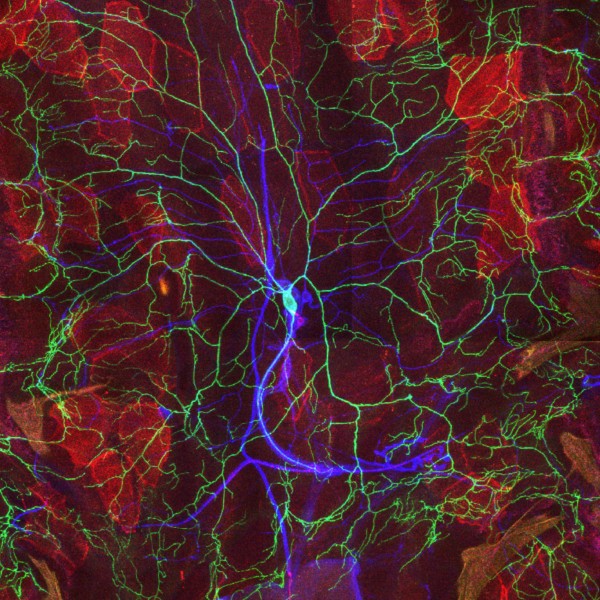
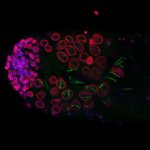
Tyrosine hydroxylase-positive neurons in tissue-cleared brain of African turquoise killifish
The killifish brain was dissected along the midline; regions enriched for dopamine- and noradrenalin-secreting neurons were retained.
The staining with anti-tyrosine hydroxylase revealed the distribution of these neuronal cell bodies and neurites in a brain fragment that was cleared with the iDISCO method.
Acquired using Olympus FV3000, resonant scanning mode using Multi-Area Time Lapse function, 30x Silicon Oil objective.
Taken by: Koh Tong Wey (KOH Tong-Wey's lab)
August 2019
Pollen tubes and callose distribution in Choysum pistil 24hours after fertilisation
The sample was stained with decolourised aniline blue and imaged with DAPI filter.
Acquired using Zeiss Axio Imager M2 at 5x magnification and stitched with ImageJ.
Taken by: Austin (Srinivasan RAMACHANDRAN's lab)
July 2019
Callose and cell wall imaging in Kailan ovules
Trial of a method for observing callose deposition in developing Kailan ovules.
Analin Blue dye stains for callose(cyan) and propidium iodide stains for cell wall(red).
Acquired using Olympus FV3000 and deconvolved through Cellsens and processed through Imaris.
Taken by: Austin (Srinivasan RAMACHANDRAN's lab)
February 2019
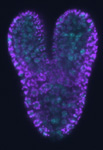
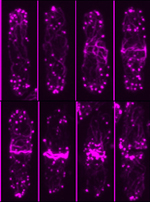
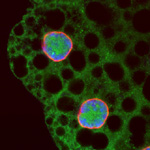
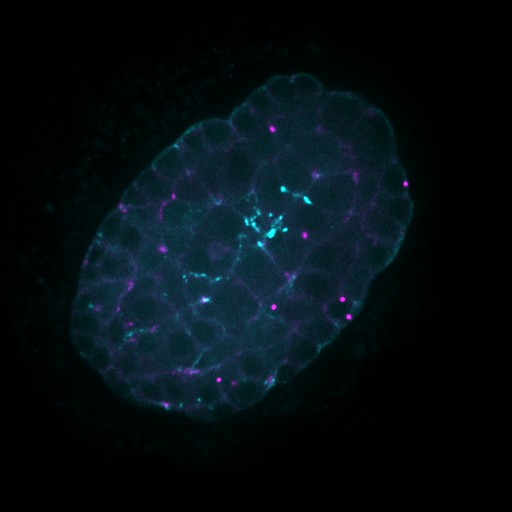
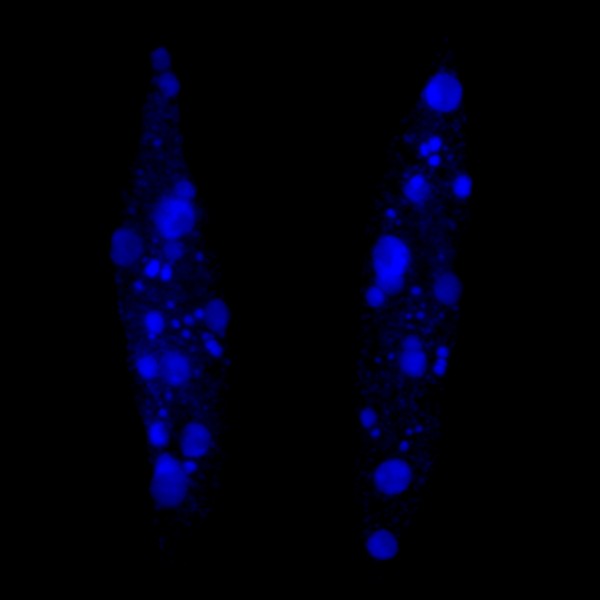
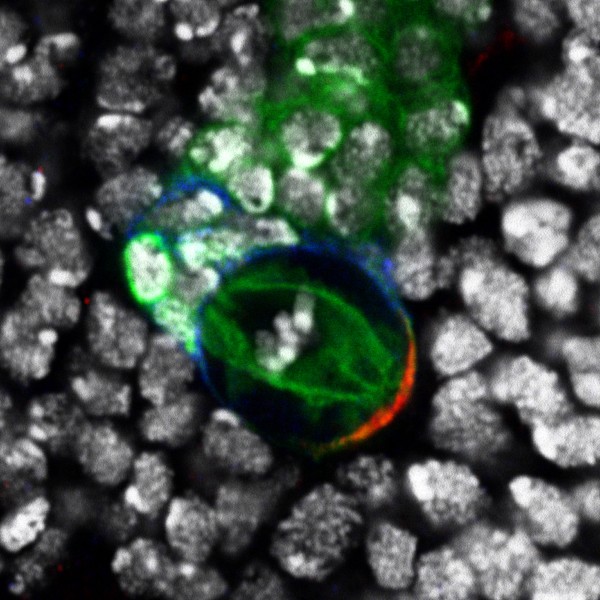
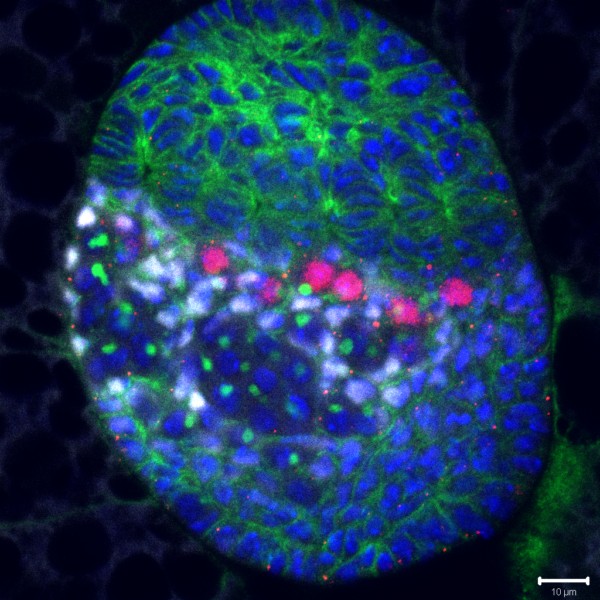
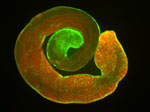
Developing Nematode Embryo
A mid-plane snapshot of developing worm embryo expressing polarity protein PAR-3(cyan) and myosin(magenta) using Nikon Ni-E motorized upright microscope fitted with a CSU-X1 spinning disk confocal system (Yokogawa Electric Corp).
Image was acquired with a Photometrics Evolve512 camera (Photometrics) controlled by Metamorph software (Intelligent Imaging Innovation Inc).
Taken by: Han ZiYin (Motegi's lab)
January 2019
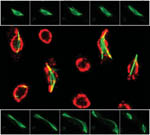
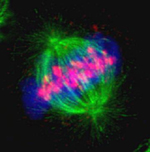
Dynamics of kinetochores (GFP-CenpA) and microtubules (mCherry-TubA) during mitosis in Magnaporthe oryzae
During mitosis, the dispersed kinetochores (GFP-CenpA) moved along the mitotic spindle (mCherry-TubA) and eventually segregated into two halves in a non-synchronous manner.
The images are maximum projections of 0.3 µm-spaced Z stacks taken on the spinning disk confocal with the Live-SR module, acquired with 100X/1.4 objective.
Taken by: Yang Fan (Naweed NAQVI's Lab)
November 2018
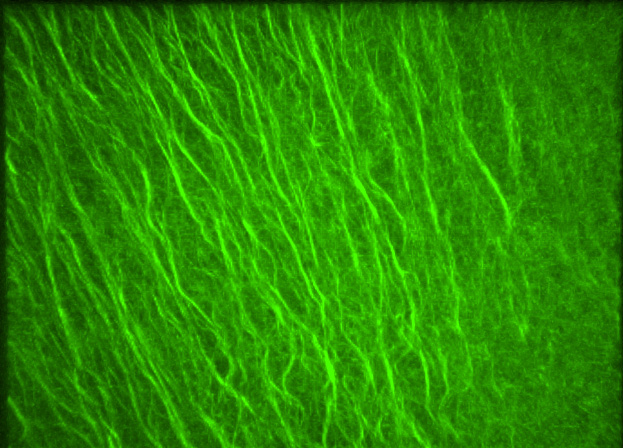
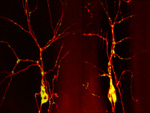
Growing microtubule in Drosophila sensory neurons
Live confocal imaging of EB1-comets movements in mutant ddaC neurons.
Embryos were manually sliced at the posterior end, and mounted with PGCs facing the coverslip.
Acquired on Olympus FV3000 Inverted
Taken by: Tang Quan(YU Fengwei's Lab)
September 2018
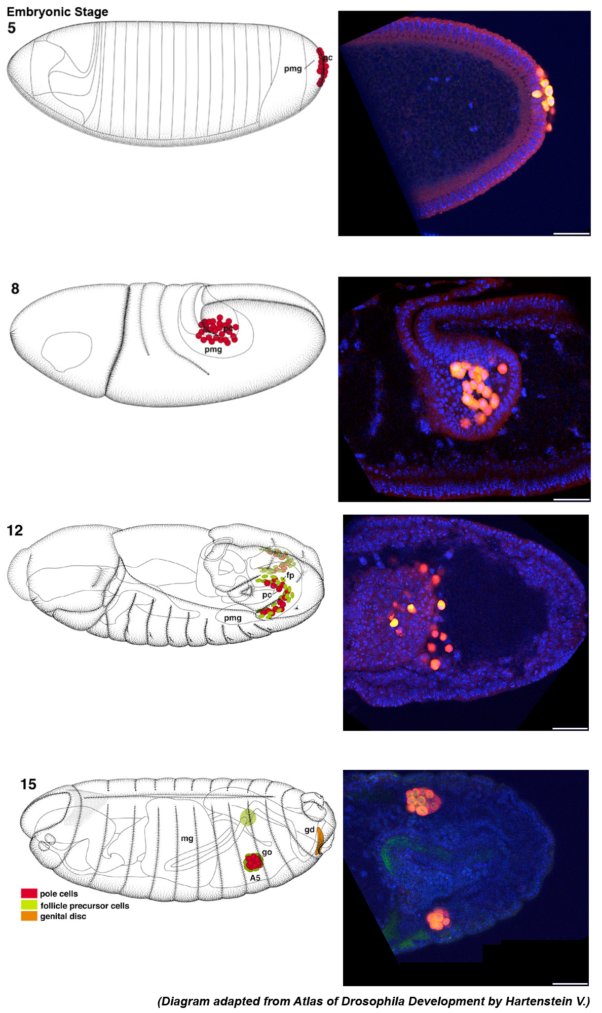
Drosophila Primordial Germ cells
Primordial Germ cells (also known as Pole cells) at the posterior pole of a Stage 5 Drosophila embryo.
Embryos were manually sliced at the posterior end, and mounted with PGCs facing the coverslip.
Green: Vasa
Red: DNA
Acquired on Leica SPEII upright, 40X lens
Taken by: Ismail Bin Osman(PEK Jun Wei's Lab)
August 2018
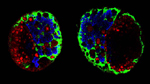
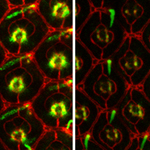
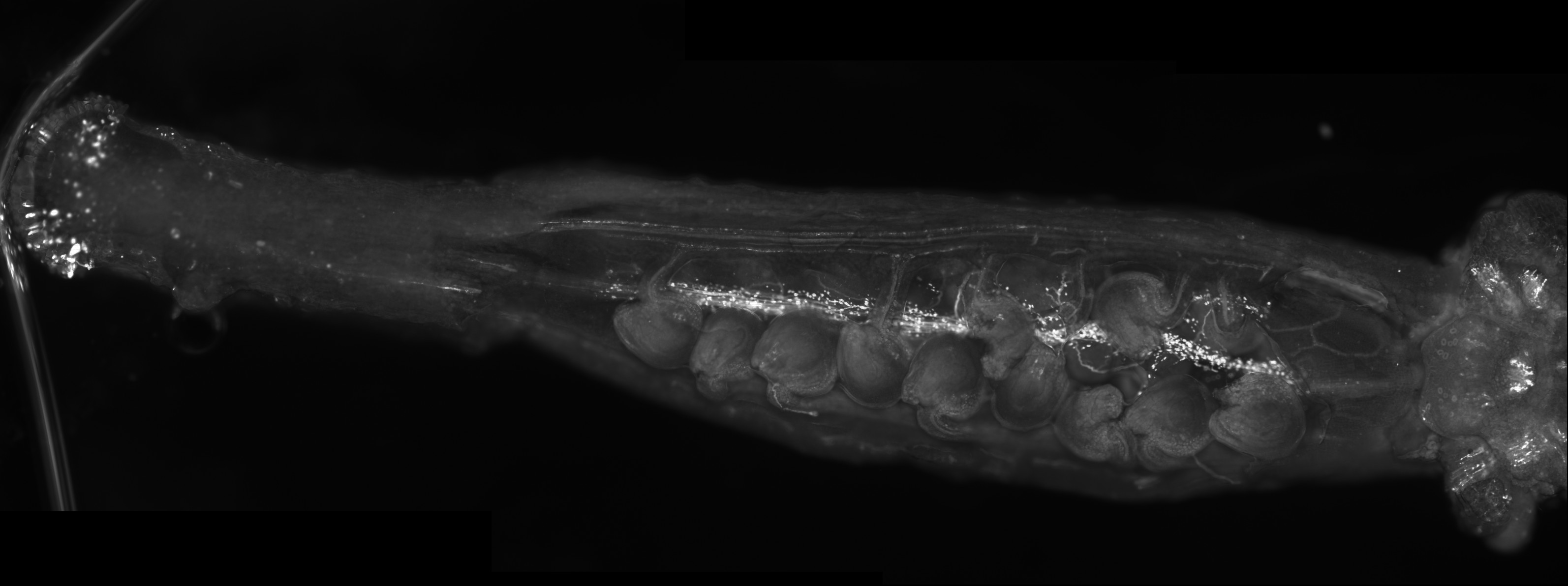
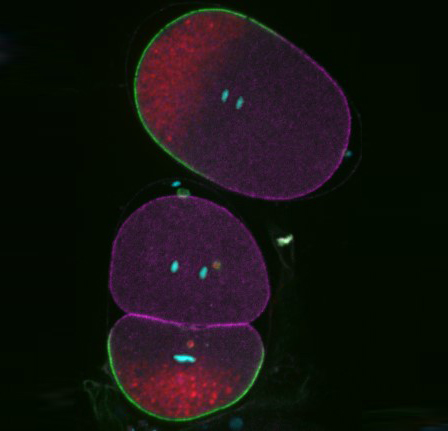
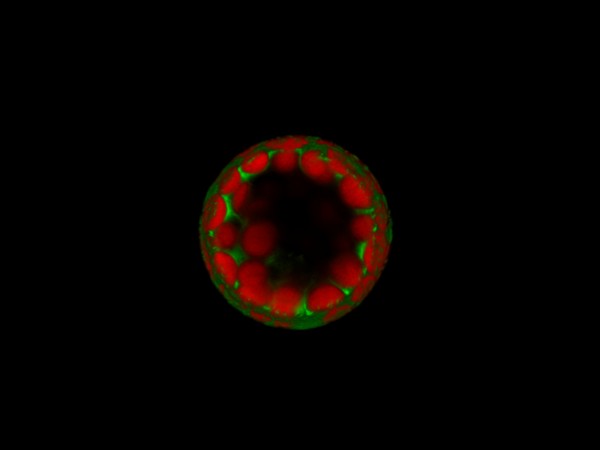
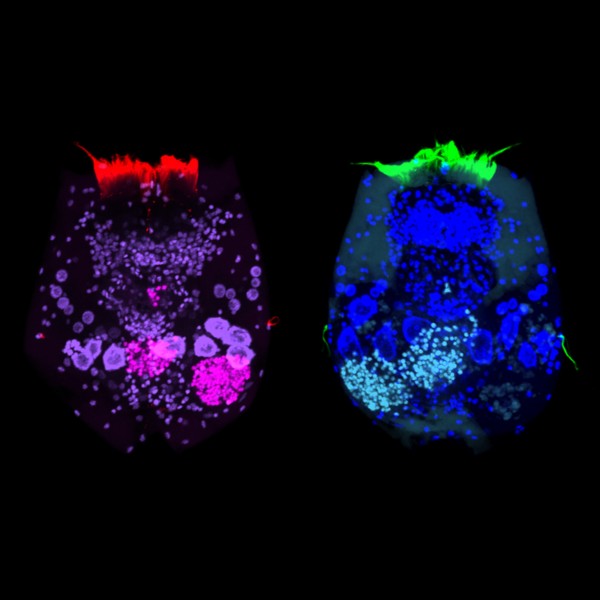
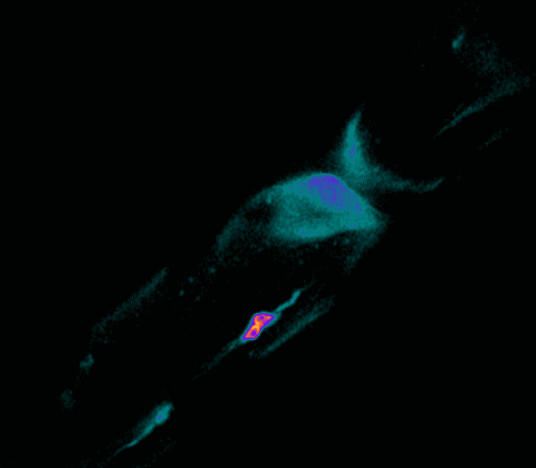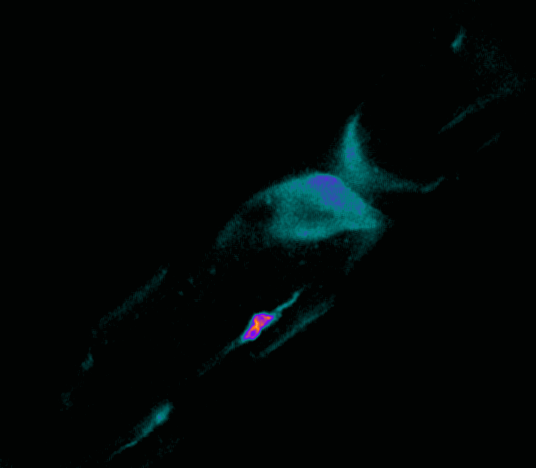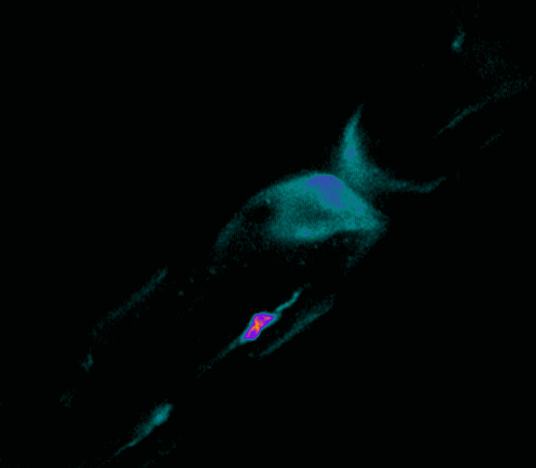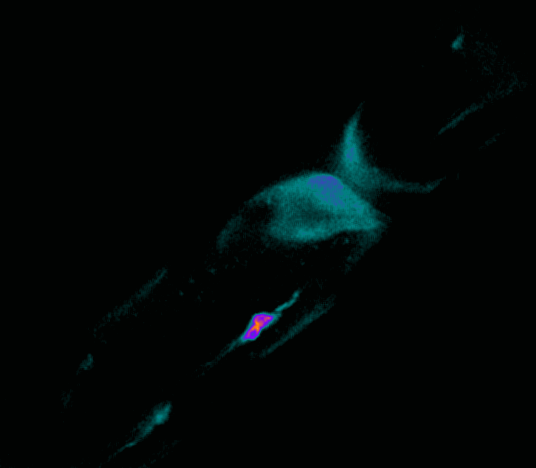
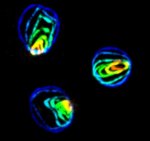
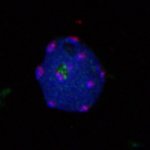
Polarisation of C. elegans Embryos
1-cell and 2-cells stage C. elegans embryos stained for PAR-6 (magenta), DNA (cyan),PAR-2 (green), and PIE-1 (red).
Embryos were observed with a CFI Plan Apochromat 100X N.A.1.45 oil immersion lens on a Nikon Ni-E motorized upright microscope (Nikon) fitted with a CSU-X1 spinning disk confocal system (Yokogawa Electric Corp).
Images were acquired with a Photometrics Evolve512 camera (Photometrics) controlled by Metamorph software (Intelligent Imaging Innovation Inc).
Taken by: Yen Wei Lim(Motegi's lab)
July 2018
Co-localization of FLAG-tagged Dsn and Disco Interacting Protein 1 (DIP1) in a Drosophila nurse cell nucleus
Drosophila labelled with FLAG-tagged Dsn(green), DIP1 protein(red) and DNA(Blue)
Acquired on SP8 system with LIGHTNING, 63X objective lens
Taken by: Tay Li Ian Mandy(PEK Jun Wei's Lab)
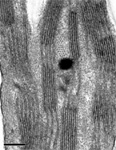
June 2018
Drosophila photoreceptor cartridge
TEM image showing 6 photorecepter terminals surrounding laminar monopolar cellsin the middle.Together,they form a cartridge.
The image was taken on the JEOL JEM1230 transmission electron microscopy.
Taken by: Claris Chong Yuin Yi(KOH Tong-Wey's Lab)
March 2018
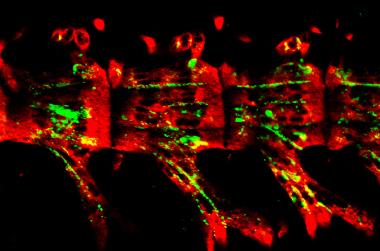
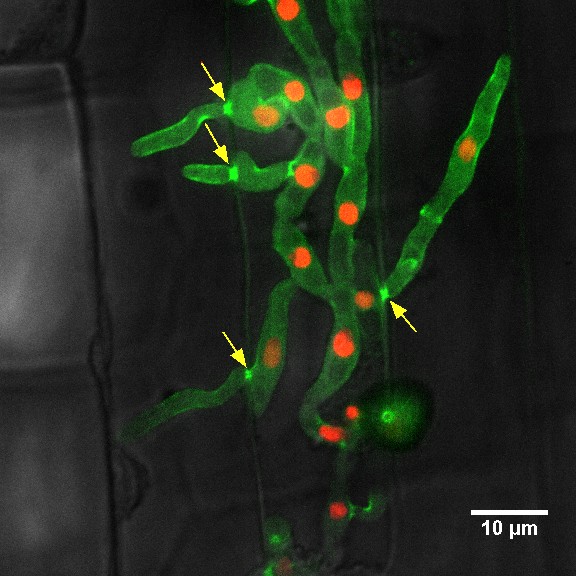
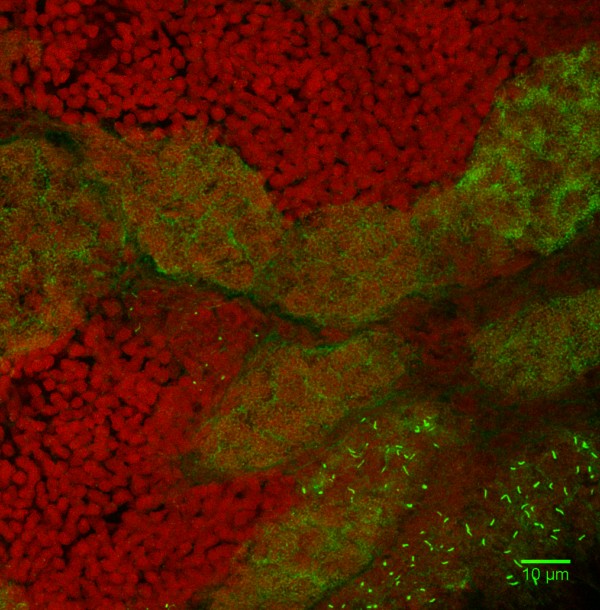
Invasive hyphal growth of Magnaporthe oryzae in live rice cells
Conidia of the M. oryzae were incubated on the rice sheath. Two days after inoculation, the fungal hyphae invaded a neighboring rice cell through plasmodesmata (arrows)
Red: nuclei (histoneH1:mCherry)
Green: septin (Sep5:GFP)
The image was taken on the spinning disk confocal, acquired with 60X/1.4 objective
Taken by: Yang Fan(Naweed NAQVI's Lab)
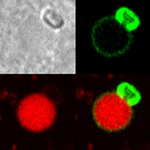
February 2018
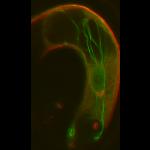
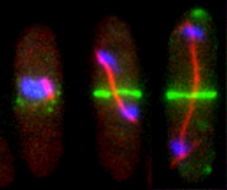
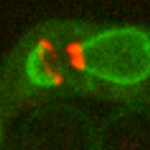
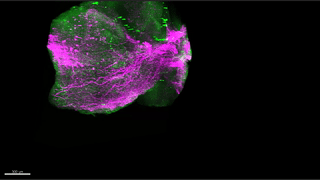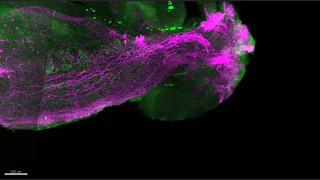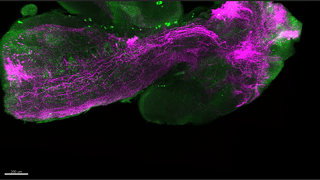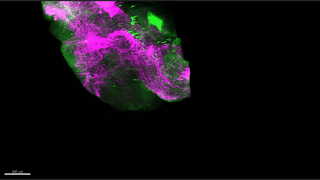
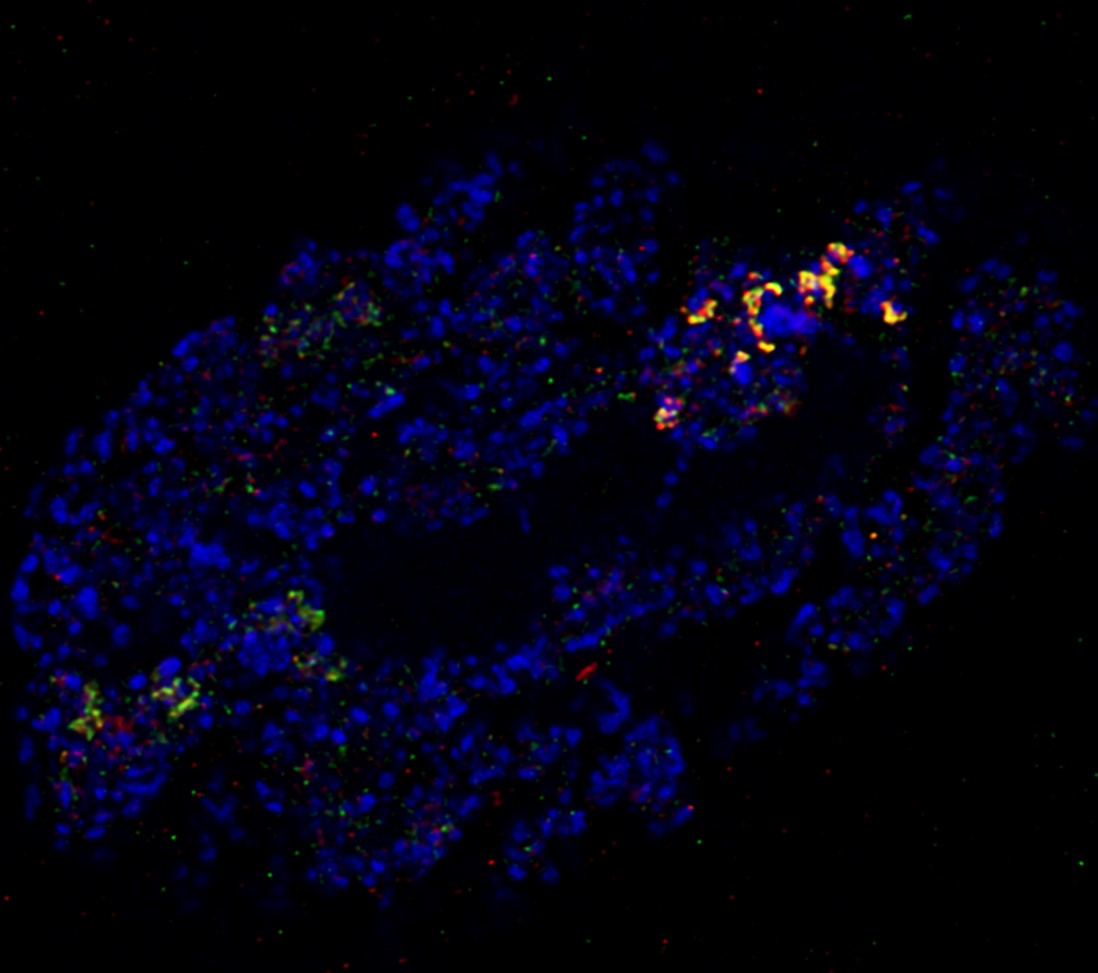
Reconstitution of C. elegans PAR polarity in budding yeast
Live budding yeast S. cerevisiae expressing C. elegans polarity proteins PAR-3(mCFP), PAR-6, PKC-3(EGFP), and PAR-2(mCherry) that spontaneously segregate into “anterior” and “posterior” domains
Acquired through SP8 with 63x objective
Image cropped with 5x digital zoom
Taken by: Han Ziyin(Fumio MOTEGI's Lab)
January 2018
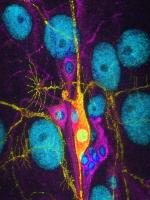
Neuronal outgrowth in N2a cells
Channels are DAPI(blue), myc-Ago(green) and dsRed(red)
Taken by: Lee Li Ming(Katsutomo OKAMURA's Lab)
December 2017
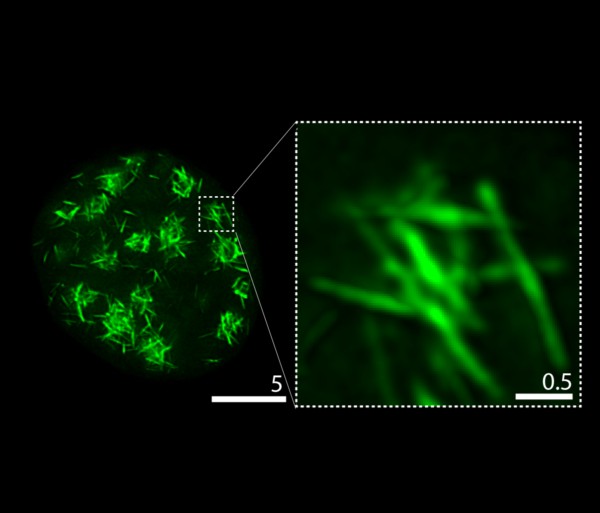
Polyampholyte Nuclear Fibrils
RD120-mGFP(Green) expressed in HeLa cells
Acquired at 100x magnification using the Leica SP8 with STED laser and deconvolved in Huygens professional
Taken by:Greig Jamie Alan (Gregory JEDD's Lab)
November 2017
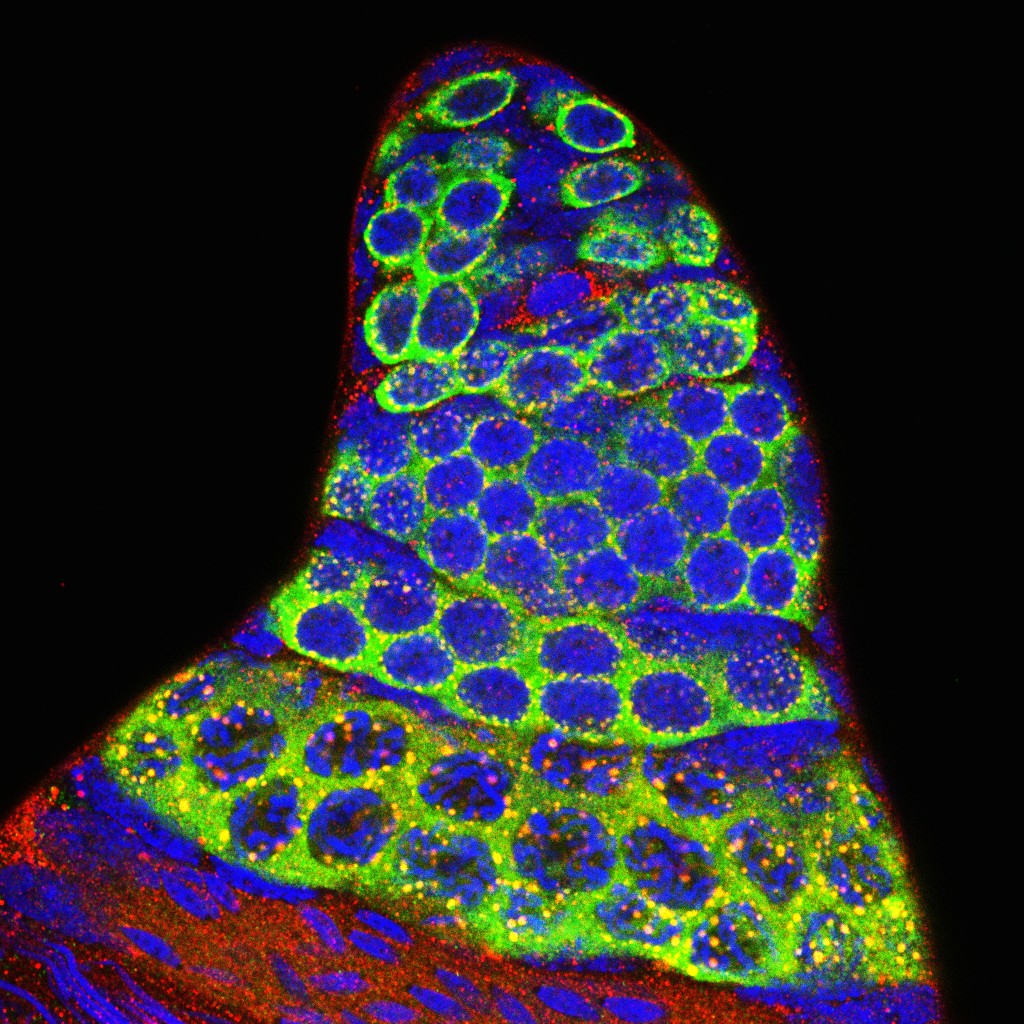
Aedes aegypti adult testis
Aedes Aegypti adult testis stained with Vasa (Germ cell marker), Nix (RNA binding protein) and Hoechst (DNA).
Green: Vasa(Ae. Aegypti) primary antibody and Alex488 secondary antibody.
Red: Nix(Ae. Aegypti) primary antibody and cy3 secondary antibody.
Blue: DNA Hoechst staining.
Image acquired using Leica SPEII upright, 40X lens
Taken by: Zhang Heng (Cai Yu's Lab)
October 2017
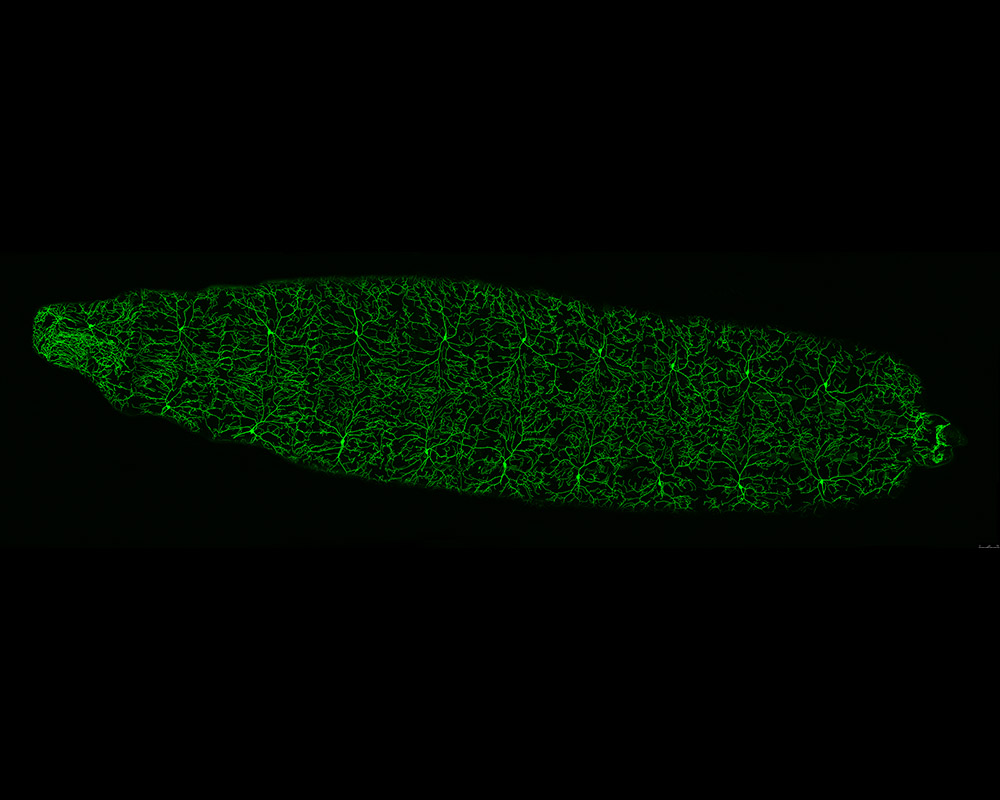
Drosophila 3rd instar Larva expressing ppk-tdGFP labelled Class IV da neurons
Image showing dorsal view of a 3rd instar larva of the fruit fly, Drosophila melanogaster.
Class IV dendritic arborization (C4da) neurons have been labelled via ppk-tdGFP construct(Green).
Time point: 96 hour after egg lay (96h AEL).
Image acquired using Leica SPEII upright, 40X zoom via tile scanning function
Taken by: Bryan Lim (Feng Wei Yu's Lab)
September 2017
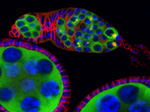
Novel nuclear bodies: Satellite bodies in Drosophila melanogaster female germline cells
A projection of a Drosophila stage 7/8 egg chamber with several gigantic nurse cells (germline cells).
Green: Novel nuclear bodies (satellite bodies) that cluster around the fourth chromosomes (dense red dots).
Red: DNA
Acquired on Exciter Upright, 40X lens
Taken by: Farzanah Akhbar (a former Temasek Poly intern) and Pek Jun Wei (PEK Jun Wei's Lab)
August 2017
Expression of Sycp3 in zebrafish testis by immunohistochemistry
Zebrafish Sycp3(Green), localizes in the spermatocytes, first detected as a small particle at preleptotene stage and becomes localized to one side of the nucleus representing meiotic bouquet structure at leptotene stage. At zygotene/pachytene stage the localization pattern is in a reticulate manner.
Nucleus is stained by DAPI(Red)
Z-stack image is Acquired on Leica SP8 Confocal Microscope,63X lens
Taken by: Jolly M. Saju ((László ORBÁN's Lab))
June 2017
Live imaging of Drosophila Larvae
Mitochondria labelled with mito-GFP(Green),actin cytoskeleton labelled with Moe-GFP(Green) and ddaC neuron labelled with RFP(Red)
Image was acquired by SPEII.
Taken by: Tang Quan (Feng Wei Yu's Lab)
April 2017
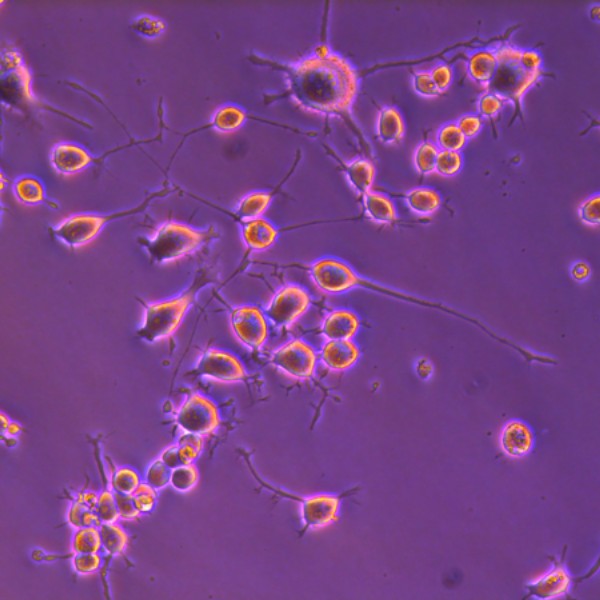
N2a differentiation into neurons
N2a cells differentiated with RA taken using phase contrast microscopy.
Image was captured by Leica DM IL LED inverted fluorescence microscope, Hiplan 1-20x objective.
Taken by: Lee Li Ming (Okamura Katsutomo's Lab)
March 2017
Peltate Glandular Trichomes on Leaf Surface
Scanning electron micrograph of spearmint leaf surface showing peltate glandular tricomes.
Image was captured by Scanning Electron Microscope Jeol JSM-6360LV
Taken by: Amarr Vaishnavi Reddy (Sarojam Rajani's Lab)
February 2017
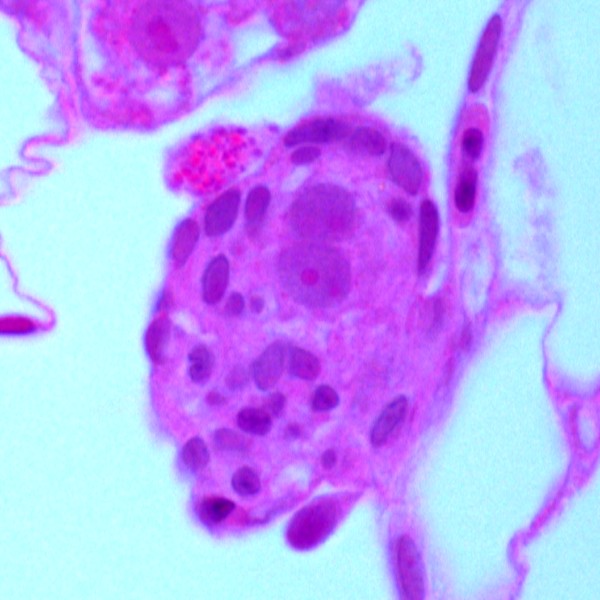
Juvenile Zebrafish Embryo
Juvenile zebrafish embryos showing early cortical alveolous stage cell. Colored by H&E stain.
Taken with Leica DFC7000T Colour Camera
Acquired on Zeiss Widefield Upright with 100x/1.4N.A. Plan-Apochromat Objective Lens
Taken by: Purushothaman. K (Laszlo Orban's Lab)
January 2017
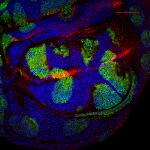
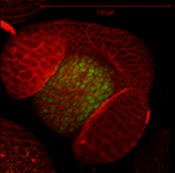
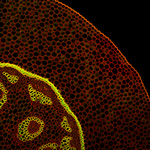
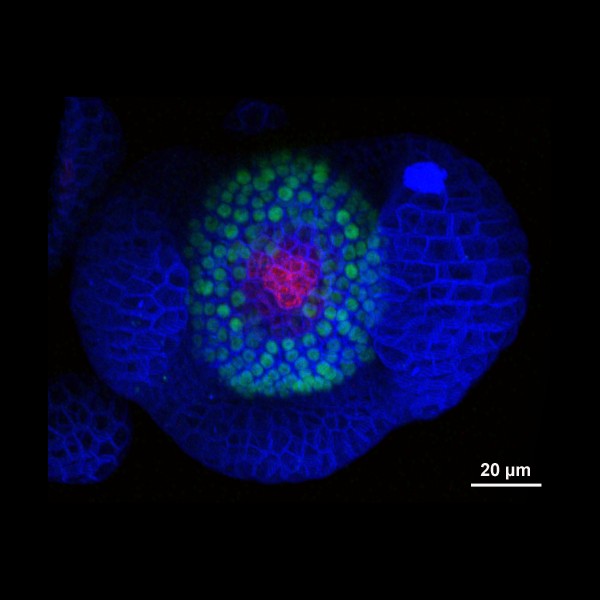
Arabidopsis Floral Bud Stage 4
Plasma membranes stained with the lipophilic dye FM4-64 (Blue). pSUP::SUP-3XVenus (Green); SUP (SUPERMAN) is a boundary gene that function to separate stamen-producing whorl 3 and carpel-producing whorl 4. pCLV3::GFP-ER (Red); a stem cell marker.
Z-Stack image rendered in 3D in Imaris software
Acquired on Leica SP8 Confocal Microscope
Taken by: Xu YiFeng (Toshiro Ito's Lab)
November 2016
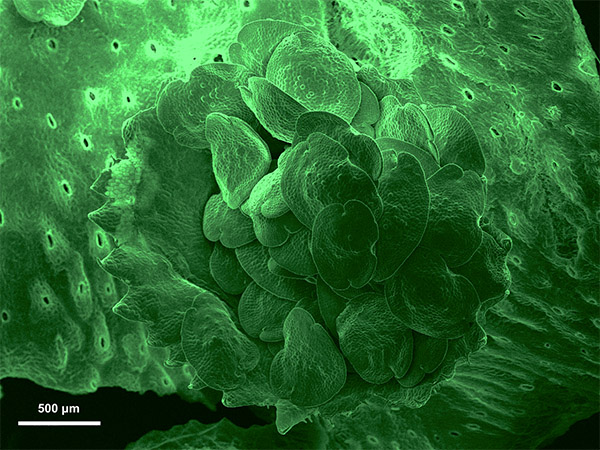
Marchantia Polymorpha Gemma Cup
Gemmae are the asexual propagules of liverworts, the oldest living land plants on earth. This mass of cells forms inside cup-like structures called gemma cups and are dispersed by rainfall to form new, genetically identical gametophytes.
Pseudo green colour added to image with Apple's Preview tool
Acquired on JEOL JSM-6360LV SEM at 35X magnification
Taken by: Sarah Seo (Urano Daisuke's Lab)
October 2016
Choy Sum (Brassica rapa var. parachinensis)
DAPI staining of an anther filament.
Maximum projection of Airyscan image done on ImageJ
Acquired on the LSM800 Airyscan Confocal Microscope with 63X Airyscan objective lens
Taken by: Norman Teo (Yu Hao's Lab)
July 2016
3D reconstituted Arabidopsis Mesophyll Protoplast
The protoplast isolated from the Arabidopsis thaliana leaves is labelled with GEF4-GFP (Green) and autofluorescence from chloroplasts (Red).
Assembled and constructed with Leica image processing software.
Acquired on the Leica SP8 Confocal Microscope with 63X Water Lens
Taken by: Zhou Zimin (Yu Hao's Lab)
May 2016
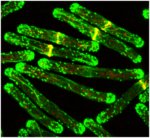
Drosophila 3rd instar larvae
3rd instar larvae expressing a transgenic PPK-tdGFP (green)
Image was stitched together from a 15x4 tile scan acquisition
Acquired on Leica SPE II
Taken by: Tang Quan (Yu Fengwei's Lab)
April 2016
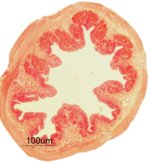
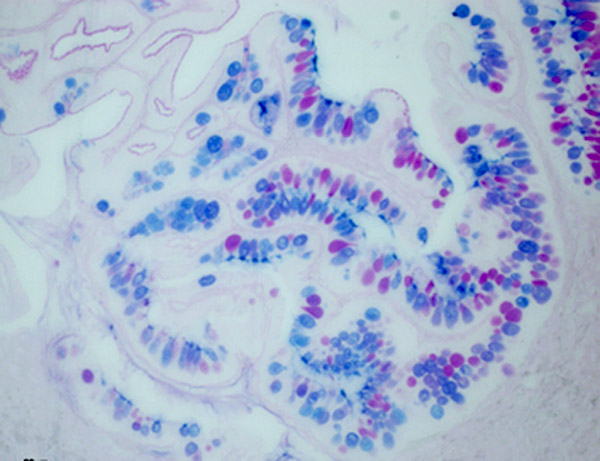
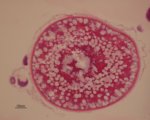
Transverse section of the Asian seabass oesophagus
Coloured with periodic acid-Schiff (PAS) and Alcian Blue (AB) stain showing neutral (Red colour in goblet cell) and acidic (Blue colour in goblet cells) glycoprotein's presents in the goblet cells.
Acquired on Zeiss Axioplan 2 mounted with a Nikon digital camera DXM 1200F.
Taken by: Purushothaman. K (Laszlo Orban's Lab).
February 2016
Wild-type L3 Drosophila brain
Antibody staining shows Dpn (Red), GFP(Green) and Ase(Blue) in wild-type L3 Drosophila brain.
Acquired on the Leica SP8 (superresolution) confocal microscope, rendered and assembled in Photoshop.
Taken by: Yang Ying (Cai Yu's Lab).
January 2016
Movie of mitotic division during appressorium development in Magnaporthe oryzae.
Conidia from the M. oryzae dual-labelled strain (CenpC-mCherry; GFP-hH1) were incubated on hydrophobic cover slips to allow appressorium development.
Mitotic division was recorded after 4 hours post-inoculation and images acquired at 20 second intervals with sequential excitation at the two wavelengths.
Centromere (red) of the nucleus (green) subsequently divided into two daughter nuclei.
Live cell imaging was performed using the spinning disk confocal, acquired with 100X/1.4 objective
Taken by: Yang fan (Naweed Naqvi's Lab).
December 2015
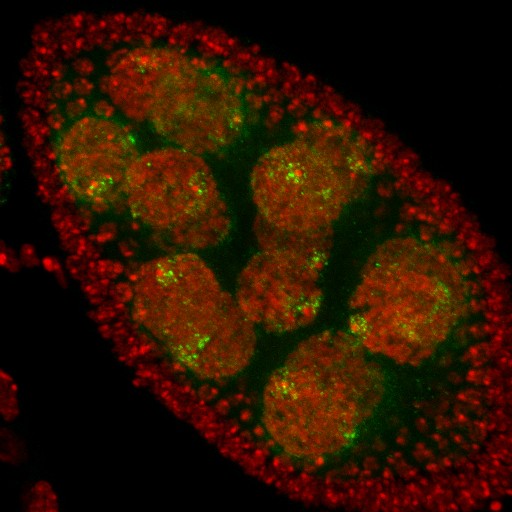
Drosophila early and middle oogenesis
Three channels stand for DAPI in Blue, F-actin in Red, a-Tubulin in Green.
Acquired on the Leica SP8 inverted confocal microscope with the 20X objective lens.
Taken by: Qin Lei (Toyama Yusuke's Lab).
October 2015
Wild type Drosophila larval sensory neurons and body wall epithelium
mCD8-RFP driven by Arm-Gal4 (Red) is expressed in Drosophila body wall epithelium. ppk-CD4-tdGFP (Green) is specifically expressed in the ddaC neuron. HRP antibody staining (Blue) shows the peripheral sensory neurons.
Acquired on the Leica SPE II upright confocal microscope, rendered and assembled in Photoshop.
Taken by: Wang Yan (Yu Fengwei's Lab).
September 2015
Wild-type L3 Drosophila brain NB in metaphase
Antibody staining shows aPKC (Red) which is enriched in the apical cortex , GFP(Green) and Mira(Blue) which is enriched in the basal cortex . DNA is dyed in white.
Acquired on the Leica SPE II upright confocal microscope, rendered and assembled in Photoshop.
Taken by: Yang Ying (Cai Yu's Lab)
July 2015
Wheel animalcules
Two rotifers fixed and stained with a-tubulin (red, green), DAPI (lavender, blue), and displaying far-red autofluorescence (magenta, cyan). Projection of 10 confocal slices.
Acquired on the Zeiss LSM 510 META inverted confocal microscope, rendered and assembled in Photoshop.
Taken by: Robyn Lim (Toshie Kai's Lab)
June 2015
Drosophila ovary at late 3rd instar larva stage
Antibody staining shows phospholyrated Mad in the precusors of germ stem cells (Red), spectromsome/fusome in all germ cells (Green) and intermingled cells (White). DNA is dyed in blue.
Acquired on the ZEISS LSM 5 exciter upright with a 40x/1.3 N.A oil objective.
Taken by: Luo Lichao (Cai Yu's Lab)
May 2015
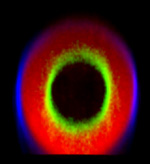
Celestial Cells
A collage of four images capturing autofluorescence from Nepenthes 'Lady Luck' pitcher plant gland cells.
Acquired on the Zeiss LSM 510 META inverted confocal microscope, rendered and assembled in Photoshop.
Taken by: Robyn Lim (Toshie Kai's Lab)
April 2015
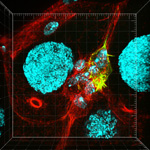
Rearranged cellular structures in dengue-infected cells
Fixed cells 24 hours post-infection; antibody staining shows viral replication centers (blue), trans-golgi network protein TGN46 (red), and exoribonuclease ERI3 (green).
Imaged on the Leica SP8 inverted confocal microscope with a 100x/1.4 NA oil objective.
Taken by: Meredith Calvert (Bioimaging and Biocomputing facility) & Alex Ward (Duke NUS)
February 2015
3D structure of Salinity Treated Mozambique Tilapia (Oreochromis mossambicus) Gills
Gills are coated with gold and imaged on the JEOL JSM-6360LV SEM with 80x magnification
Taken by: Purushothaman. K (Lazlo Orban’s lab),
Wang Jing Fang &
Ouyang Xue Zhi
(Bioimaging and Biocomputing facility)
January 2015
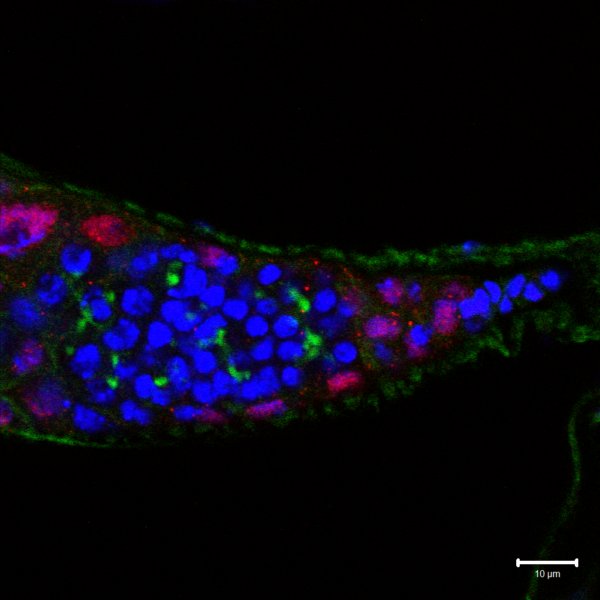
Drosophila Embryo Germline Development
The development of drosophila germline in embryo from pole cells to progenitor germ cells and the formation of gonads
DNA stained blue, germline specific gene in green and vas in red.
Taken on Zeiss Exciter upright, Plan-Neofluar 20x/0.5 objective
Taken by: Ryan Teo Yee Wei (Toshie Kai’s lab)
October 2014
Germarium Carrying Pre-diferrentiated Germ Cells
Fusome, is stained in green. pMad (Red) is positive in escort cells. DNA dyes in blue.
Taken on the Zeiss 510 META upright confocal with the Plan-Neofluar 40X/0.75 objective
Taken by: Luo Li Chao (Cai Yu’s lab)
September 2014
Response of ppk23 gustatory neurons on the foreleg of a male Drosophila melanogaster fly to the courtship pheromone CH503
ppk23 neurons were labelled with the genetically encoded calcium indicator GCaMP5.
Time series of Z-stacks captured on the MicroLambda spinning disk confocal.
Taken using the 60x objective and the ORCA-Flash 4.0 camera
Taken by: Shruti Shankar (Joanne Yew’s lab)
August 2014
Microtubule staining for stage 4 Drosophila egg chamber
Stage 2 and stage 4 Drosophila egg chambers stained with anti-tubulin GFP and DAPI for cellular nuclei
Image was taken on the Leica SP8 with the 63x Oil Objective
Taken by: Qin Lei (Toyama Yusuke’s lab)
July 2014
Nuclear division during appressorium form
(of Magnaporthe oryzae)
A dual-labeled M. oryzae strain expressing a nuclear mCherry and a cytoplasmic GFP during appressorium formation.
See the cover of July's Fungal Genetics and Biology 68: 71-76. (here) for more images of different fluorescent-labeled M. oryzae strains!!
Taken by: Yang Fan (Naweed’s lab)
May 2014
Bristle in the Wing
Cross section of a wing of a butterfly. Image was capture using the JEOL 1230 Transmission electron microscope
Taken by: Dr Dion Emilie Julie Pauline (Joanne Yew's Lab)
April 2014
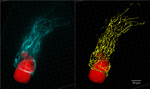
Endosomal Landscapes
The late endosomes (Rab7-GFP) scaffold the active G proteins (Rgs1-mCherry). Rgs1 does not associate with the early endosomes (Rab-5-GFP).Image was taken on the Microlambda Spinning disk with a 100x/1.4 oil lens and the image is a 3D rendering of five 0.5um slices.
Taken by: Ravikrishna Ramanujam (Naweed's Lab) and Meredith Calvert (Bioimaging and Biocomputing facility)
March 2014
Drosophila adult brain whole mount
Here is an image of a Drosophila brain showing the neurons, ovarian tumour and numerous nuclei. Image was acquired on the Zeiss Exciter upright confocal microscope with a 20x objective lens.
Taken by: Ryan Teo Yee Wei (Toshie Kai's Lab)
February 2014
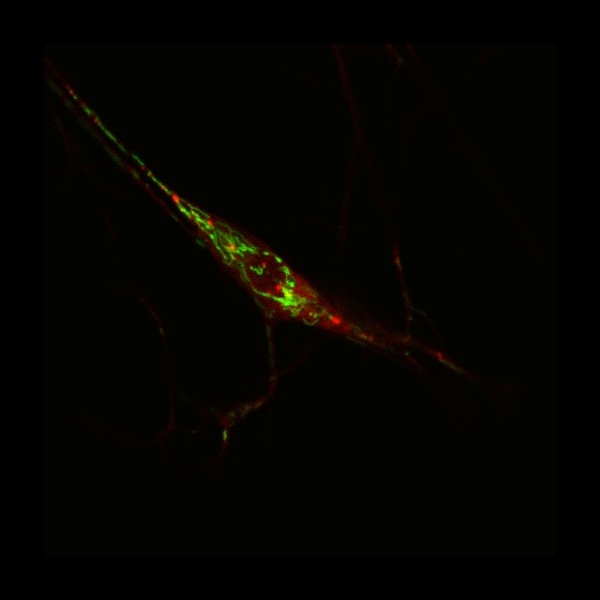
Larva's neurons
The ddaC neurons of the Drosophila melanogaster larvae labelled with ppk-TDGFP. Images were taken at 20x resolution with Leica SPE
Taken by: Jack Wong (Fengwei's Lab)
January 2014
Histology of Carp Ovary
Coloured by Haematoxylin and Eosin stain. Taken with a 10x objective lens
Taken by: Shubha Vij, Doreen & Purushothaman (Laszlo Orban's Lab)
December 2013
Arteflake
A composite of four images of Artemia salina (brine shrimp) nauplii made into the shape of a snowflake. E. coli expressing eGFP and mCherry fluorescent proteins (blue) are present in the gut of the nauplii. Acquired on the Zeiss Axioplan 2 upright widefield fluorescence microscope, rendered and assembled in Photoshop.
Taken by: Robyn Lim (Dr.Toshie Kai's Lab)
November 2013
Contractile ring contraction in vitro
Rings of fission yeast cell ghosts were visualized by RIc1(myosin light chain)-3xGFP using a 100x objective lens.The images of the same ring captured every 20 seconds were merged in rainbow colors. Blue indicates the time of ATP addition.
Taken by: Mishra Mithilesh (Mohan Balasubramanian's lab)
October 2013
Happy Halloween!!
A spooky mutant Arabidopsis inflorescence displays abnormal flowers and demonstrates some characteristics of secondary inflorescence shoots.
Image was acquired using the JEOL JSM-6360LV Scanning Electron microscope.
Taken by: Norman Teo (Dr. Yu Hao's lab)
September 2013
Mutant Drosophila testis
Showing stellate protein in the primary spermatocytes, perinuclear lamina and DNA. The image was acquired on the Zeiss LSM 510 META inverted, with the 40x objective.
Taken by: Emilie Quenerch'du (Toshie Kai's lab)
July 2013
Histology of Asian Seabass ovary
It is colored by Haematoxylin and Eosin stain. Taken with a 100x objective oil immersion lens.
Taken by: Purushothaman Kathiresan and Inna Kuznetcova
(Laszlo Orban's lab).
June 2013
Arabidopsis leaf nuclei
Arabidopsis leaf nuclei, showing DAPI-stained DNA (blue), immunostaining for H3K9me2 and ATRX-GFP. The image is a single slice captured using the Zeiss Exciter upright confocal microscope with 40x magnification and zoomed in 6x.
Taken by: Wang Haifeng (Fred Berger's lab).
May 2013
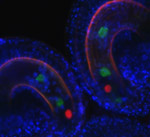
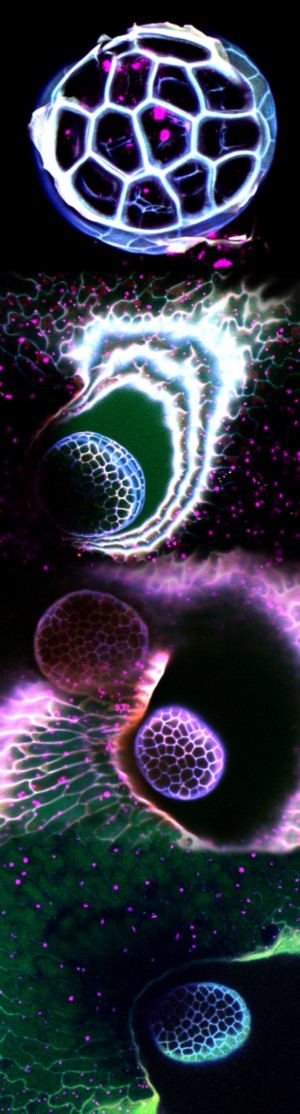
The Nuclear Comet
Arabidopsis ovule showing Central cell Lifeact-Venus, Egg cell H2B-mRFP and Synergid cell mCherry; surface rendering by Imaris. Stack of 30 slices acquired on the MicroLambda spinning disk, 60X oil lens.
Taken by: Tomokazu Kawashima (Dr. Fred Berger's lab)
April 2013
What's in a Pitcher?
Mosqito larvae from the pitfall trap liquid of the pitcher plant Nepenthes rafflesiana. Image was acquired using the JEOL JSM-6360LV Scanning Electron microscope.
Taken by: Leonora Bittleston (Pierce & Pringle Labs, Harvard University) and Fiona Chia (TLL Bioimaging & Biocomputing Facility)
February 2013
Happy Year of the Snake!!
Drosophila testis stained with Anti-alpha-spectrin, Anti-aubergine. Overlaid images were acquired using Zeiss Widefield Upright microscope with a CoolSnap CF camera and a 10x lens.
Taken by: Ryan Teo Yee Wei (Dr.Toshie Kai's Lab).
January 2013
Fission yeast cdc25-22 mutant cells.
Lifeact-mCherry, Cdc12p-3Venus. Single wavelength images were acquired on the MicroLAMBDA spinning disk with a 100x oil lens; maximum intensity projection of 11 axial slices.
Taken by: Jacky Junqui Huang(Dr. Mohan Balasubramanian's lab)
December 2012
Merry Christmas Chromosomes
Chromosome from Drosophila larval salivary gland. Anti-pol II, DNA, anti-H3K9Me3. Anti-Stellate protein star- adult testis from a piRNA mutant, tejas. Images were acquired on LeicaSPE confocal, 63x lens.
Taken by:Sophie Hamel (Dr.Toshie Kai's lab), Meredith Calvert (Bioimaging and Biocomputing Facility)
November 2012
Andy Warhol's Histoblast Print
Drosophila pupal epidermis. DE-Cadherin::mTomato (upper left); ECFP-Venus FRET pair (upper right) ECFP (lower left) Venus (lower right). Images were acquired on Zeiss LSM 510 META inverted confocal with 40X water lens.
Taken by: Teng Xiang (Dr. Yusuke Toyama's lab)
October 2012
Drosophila testis
Drosophila testis showing Krimper, RNA Polymerase II and DAPI. The image was acquired on the Zeiss Exciter upright confocal microscope with the 40X lens.
Taken by: Chang Guang Yu Jacintha (Toshie Kai's lab).
September 2012
Esophagus of seabass fish
Tissue section from the esophagus of 1 month seabass fish. The tissue has been stained with H/E and captured using 10 X magnification.
Taken by:Purusothaman Kathiresan (Dr. Laszlo Orban's lab)
August 2012
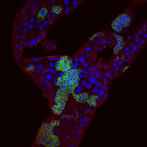
Superresolution Microscopy: Drosophila ovary
Antibody staining for Lamin, Kumo and Krimper. Captured on the Zeiss ELYRA Superresolution system.
Taken by: Meredith Calvert (Bioimaging and Biocomputing Facility) and Amit Anand (Toshie Kai's lab)
June 2012
Wts mutant wing disc in Drosophila
Shows Wts mutant clones, Wg protein staining and DNA in blue. Scale bar is 10 μm.
Taken by: Liu Zhong (Dr. Cai Yu's lab)
May 2012
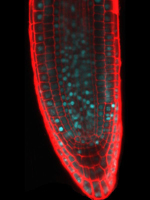
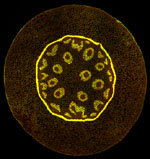
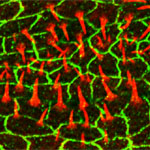
Lateral root initiation site in the Arabidopsis root.
The primary root was labeled by Propidium iodide while the lateral root cells were labeled by pLBD16P::TUA6-GFP.
Taken by:Zhou Zimin (Dr. Tongda Xu's lab)
April 2012
Karyogamy in Arabidopsis.
F-actin pFWA::LifeAct-Venus, cytoplasm pFWA::mCherry and sperm cell nuclei pHTR10::H2B-mRFP in the central cell.
Taken by: Tomokazu Kawashima (Dr. Fred Berger's lab)
March 2012
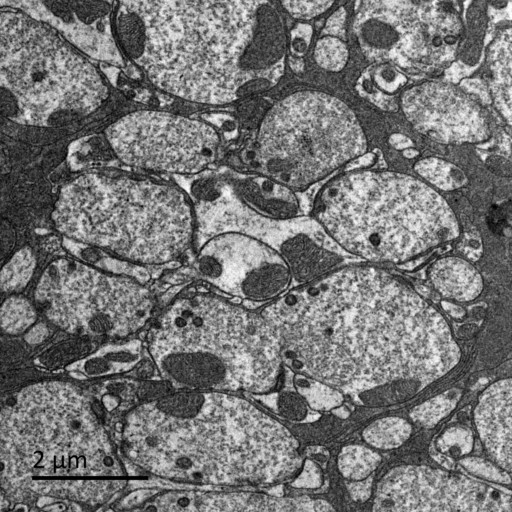
Magnaporthe mycelia
Magnaporthe myceliaunder nitrogen starvation, visualized by TEM. Autophagic bodies accumulated in the vacuole.
Taken by: Deng Yi Zhen (Dr. Naweed Naqvi's lab)
February 2012
Notch mutant cells in Malpighian tubules (MTs) of Drosophila melanogaster.
MARCM clones , Delta protein stained with CY3 and DNA stained with Topro 3.
Taken by: Liu Sen (Dr. Yu Cai's lab)
January 2012
Deformation of fission yeast nuclear envelope
Deformation of fission yeast nuclear envelope by Ba-TubZ polymers during mitosis in fission yeast.
Taken by: Dr. Mithilesh Mishra (Mohan Balasubramanian's Lab)
December 2011
Mint leaf
Scanning electron micrograph of a mint leaf (Mentha spicata) showing the 3 types of trichomes present on it.
Taken by: Dr. Sarojam Rajani
November 2011
Arabidopsis thaliana flower bud
Expression of Agamous in Arabidopsis thaliana flower bud stage 4. AG::GFP and cell membrane stained with FM4-64
Taken by: Wee Wan Yi (Toshiro Ito's Lab )
October 2011
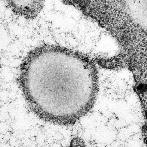
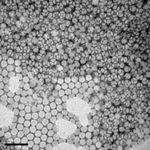
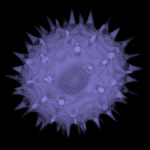
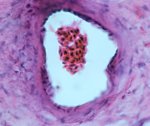
Enterovirus 71
Enterovirus 71, visualized by TEM. The icosahedral architecture of this 30nm virus is reflected in the hexagonal shapes, negatively stained using phospho-tungstic acid.
Taken by: Tanja Kiener(Jimmy Kwang's Lab ).
September 2011
Fission yeast (S. pombe)
Fission yeast (S. pombe) expressing rlc1-3GFP (Regulatory light chain of Myosin2) withmCherry-atb2 (Tubulin) and DAPI to visualize the nucleus.
Taken by: Dhivya Subramanian (Mohan Balasubramanian's lab)
August 2011
Drosophila germ-line cell
FACS-sorted Drosophila germ-line cell expressing Vasa and transgenicVasa-GFP. The spectrosome is labeled with alpha-Spectrin antibody.
Taken by: Robyn Lim (Toshie Kai's lab)
July 2011
Drosophila testis
Drosophila testis showing the expression of the PIWI-family protein Aubergine, Vasa, and costaining for DNA.
Taken by: Emilie Quénerch'du (Toshie Kai's lab)
April 2011
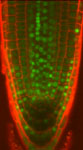
Arabadopsis root tip
Arabadopsis root tip showing nuclei expressing methyl transferase-CFP and cell boundaries stained with propidium iodide.
Taken by: Pauline Jullien (Fred Berger's lab ).
Drosophila pupa body wall
Drosophila pupa body wall preparation.Shows nuclei with Ecdysone Receptor B1, Mical is a cytoskeletal molecule expressed throughout the neurons andPickpocket-gal4 driven UAS-GFP.
Taken by: Daniel Kirilly (Yu Fengwei's lab)
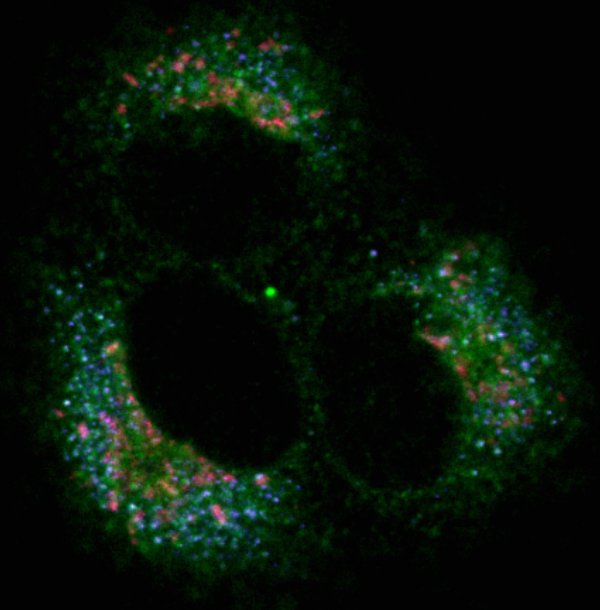
HeLa cell
HeLa cell fixed during metaphase triple lableled for Aurora B, microtubules and DNA. Image acquired on the Zeiss LSM 510 META inverted confocal and processed using the Imaris software.
Taken by: Vinayaka Srinivas (Maki Murata-Hori's lab)
Microtubules in Zebrafish
Parallel arrays of microtubules at the vegetal cortex of the zebrafish embryo in early development.
Taken by: Tran Duc Long (Karuna Sampath's lab)
Embryo of Arabidopsis thaliana
A confocal image of a torpedo stage embryo of Arabidopsis thaliana with a chromatin CFP marker and background autofluoresence. Image acquired using the Zeiss LSM 510.
Taken by: Pauline Jullien (Fred Berger's lab)
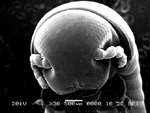
A millipede's head
Acquired using the SEM.
Taken by: Fiona Chia (Bioimaging and Biocomputing facility)
Two brain lobes of the Drosophila melanogaster 3rd instar larval brain.
The neuroblasts are labelled with Deadpan antibody (red), while the glial cells are marked with GFP (green). The differentiated neurons are labelled with Elav antibody (blue).
Taken by: Serene Tan & Woo Mun Wah (Cai Yu's lab)
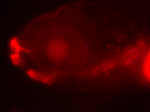
Zebrafish
A lateral view of a 6 day old zebrafish with a trasposon insertion expressing an mCherry reporter in the mouth/anterior gut, lens and some head muscles.
Taken by: Helen Quach Bao Ngoc & Karuna Sampath
Neurospora crassa septation
Actin is labelled with Lifeact-GFP (green), the plasma membrane with FM4-64 (red) and the cell wall with calcofluor (blue).
Taken by: Meredith Calvert (Biocomputing and Bioimaging facility) & Graham Wright
Contractile actomyosin ring formation
Montage of contractile actomyosin ring formation in Schizosaccharyomyces pombe (fission yeast). Actin patches and cables are stained with phalloidin in fixed cells.
Taken by: Yinyi Huang (Mohan Balasubramanian’s lab).
An autofluorescent plant vascular bundle imaged on a Zeiss 510 Meta in 2 channels
Taken by: Graham Wright
Section of a Drosophila ovary (germarium)
Taken by: Daniel Kirilly
Mutant follicle cells (green) expanding on the surface of fruit fly egg chamber
Taken by: Daniel Kirilly
Invitrogen Molecular Probes' Fluocells slide #1
Imaged on a Zeiss Exciter in 3 channels - DAPI, Alexa Fluor 488 phalloidin and Mitotracker Red CMXRos
Taken by: Graham Wright
"Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting"
Fangfang Liu et al. (2008) Journal of Cell Biology, 180: 325-339 (Greg Jedd's lab)
An autofluorescent transverse section of Convallaria sp. imaged on a Zeiss Exciter in 2 channels
Taken by: Graham Wright
An autofluorescent plant vascular bundle imaged on a Leica SP5 in 3 channels then rendered in Imaris
Taken by: Graham Wright
A border cell cluster in a Drosophilaegg chamber. F-actin (red), DNA (blue) and Moesin-GFP in one cell of the cluster (green).
Captured using the Leica SP5. De-convolved using Autoquant and 3D rendered in Imaris.
Taken by: Adam Cliffe
A zebrafish brain (3-5 day old fish), triple labelled for nuclei (blue), acetylated tubulin (axons, green), and EphB receptors (red).
The image also featured in an exhibit entitled "Neuroscapes: An Exhibit in Honor of Santiago Ramon y Cajal" and in a related book.
Taken by: Michael Hendricks
Peripheral neurons (blue and green) and the nuclear ecdysone receptor (red) in a Drosophila larva.
Taken by: Daniel Kirilly
A DIC image of a freshly harvested human cheek cell (Graham's).
Image acquired using a Zeiss LSM 5 Exciter, 40x 1.3 NA objective.
Taken by: Graham Wright
A confocal image of a root tip ofArabidopsis thaliana with a chromatin GFP marker (green) and propidium iodide counterstaining (red).
Image acquired using the Zeiss 510 META, inverted.
Takeb by: Mathieu Ingouff
An autofluorescent transverse section of Convallaria sp.
Imaged on a Zeiss LSM 510 in 2 channels. This composite of 12 images was captured using the motorized stage then receonstructed using ImageJ'sMosaicJ plugin
Taken by: Graham Wright
The Drosophila embryonic muscles (red) with the Rolling pebbles protein (green) localized along the surface of muscles where they adhere to one another.
Images were acquired using the Zeiss Meta 510 upright confocal.
Taken by: Sree Devi Menon
A montage of two pairs of Drosophilaperipheral neurons labeled with RFP.
Mitochondria are shown in green due to the presence of mito-GFP.
Takeb by: Daniel Kirilly
An Arabidopsis thaliana root tip was stained with FM4-64, imaged in xy and z on a Zeiss LSM 510, then 3D and iso-surface rendered using Imaris.
Taken by: Graham Wright
Microtubules in the NRK (normal rat kidney epithilia) cells by immunofluorescence staining.
A Z-stack was captured using the Zeiss LSM 510 META inverted confocal then projected.
Taken by: Zhu Xiao Dong
Trichomes (hairs) on the ventral surface of a Drosophila embryo which is mutant for wingless.
The main component of the hair is actin, here labeled with Cy3 (red) coupled phalloidin. Green, anit-beta-catenin staining, shows the cell membranes.
Taken by: Jacqueline Chin & Marita Buescher
2008
Inside a Winter Jasmine flower bud.
Z-series of images acquired using the spinning disk confocal then the 3D volume rendered and animated using the Imaris software.
Taken by: Graham Wright
3D projection of a pollen grain, imaged on the Zeiss LSM 5 Exciter upright
Taken by: Cristiana Barzaghi
Two Arabidopsis ovules side by side that have been fertilized by pollen containing HTR12 histones labeled with GFP (green).
Male HTR12 histones are expressed in the endosperm nuclei, of which the centromeres are distinguished as dots. The egg cell is labeled in red with RFP and autofluorescence of the teguments is visualized in blue.
Taken by: Sarah Holec & Fred Berger
3D surface rendering of an enucleated mammalian erythroblast.
The extruded nucleus is visualized by Hoechst staining (blue), while the organization of F-actin and spectrin in the incipient reticulocyte are depicted in red and green, respectively. Z-stacks acquired using the Zeiss 510 META inverted. 3D reconstructed using Imaris software.
Taken by: Tzutzuy Ramirez Hernande
A Drosophila melanogaster brain stained with anti-Repo antibody labelling all glia (red) and a GFP reporter line labelling a subset of glia which form the blood-brain barrier surrounding the brain (green).
Taken by: Jacqueline Chin & Marita Buescher
The tubular mitochondrial distribution (tagged with GFP) in invasive hyphae of Magnaporthe grisea (within the host rice tissue - red).
The circled part shows its spread from one host cell to another through plasmodesma. The fungal hyphae are ~ 1-2 μm in diameter.
Taken by: Archna Patkar & Rajesh Patkar (Naweed Naqvi's lab)
The fission yeast,Schizosaccharomyces pombe, during meiosis II.
meu14-mCherry (leading edge protein; red); gfp-psy1(forespore membrane protein; green). This cell is a mutant in which two of the four forespore membranes can form and encapsulate the haploid nuclei, but two are defective and form a cup-like structure incapable of encapsulating the nuclei.
Taken by: Yan Hongyan (Mohan's lab)
The developing eye imaginal disk of Drosophila melanogaster, ~3 days before eclosion.
The left panel is wild-type, the right panel is a mutant in which most of the bristles, hair-like sensory organs, are missing. Red: cadherin; green: 22C10, which stains bristels.
Taken by: Valérie Hilgers (Cohen lab) & Smitha Vishnu (Rørth lab)
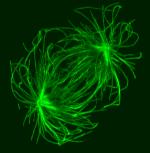
Rhodamine-labelled Taxol-stabilized microtubules bundled in vitro by purified recombinant fission yeast Transforming Acidic Coiled-Coil protein Mia1p.
Taken by: Rahul Thadani (Oliferenko lab)
.png)




-9Jan2017-Cer-yoPAR-6-PKC-3-Cer-mCherry-PAR-2-EGFP-PAR-3WT-col2-live-Series003-merge .jpg)
.jpg)


_thm.jpg)